Research Article - Biomedical Research (2022) Volume 33, Issue 7
Molecular docking of a beta-amyloid protein involved in Alzheimer?s disease with Thioflavin-T as a ligand.
Parul Singh, Neha Sharma*, Bharti Choudhary
Department of Biotechnology, College of Engineering and Technology, IILM University, Greater Noida 201301, Uttar Pradesh, India
- Corresponding Author:
- Neha Sharma
Department of Biotechnology
College of Engineering and Technology
IILM University
Greater Noida 201301
Uttar Pradesh
India
Accepted date: 07 September, 2022
Abstract
Molecular docking provides tremendous useful scope for drug design and drug discovery. Molecular docking has a use in the therapeutic industry which enables us to know the interaction between ligand and protein. Also, it gives information regarding the binding site of protein where the ligand binds. Structural details, binding energy, and activation energy of chemical compounds are determined with the help of docking. In this study, the Autodock Vina tool with PyMOL visualizer is used to perform the docking between Beta-amyloid protein and Thioflavin-T ligand. This research offers the development of a high throughput method in biology and chemistry that can be used as bioinformatics tools for drug design. This method is based on molecular docking which is performed using the Windows platform. After performing molecular docking total of nine poses are generated this shows the different binding sites of protein-ligand interaction.
Keywords
Autodock Vina, Drug design, Protein-ligand, Beta-amyloid, Thioflavin-T, Molecular docking.
Introduction
One of the most important and widely used tools for drug discovery is molecular docking. The demand for new therapeutic drugs has been widely increased these last few years after the finalization of the human genome project. During this time techniques like nuclear magnetic resonance spectroscopy, crystallography, and protein purification have been developed and used in many structural details of different proteins. The complex structure of ligand protein can be predicted using a computational method such as molecular docking. It is one of the most preferable methods for drug discovery [1,2].
High throughput screening is one of the traditional methods and virtual screening is the new advanced and direct method. It is of low cost and is very efficient in screening [3,4].
There are two types of virtual screening that is ligandbased and structure-based. Ligand-based methods like pharmacophore modeling and Quantitative Structure- Activity Relationship (QSAR) are used when there is no prior knowledge of the structure and active sites of ligands. As far as molecular docking is concerned structure-based method is used. This method is used since the early 1980’s [5].
In pharmaceutical research molecular docking is an important tool in which programs are based on various algorithms. In the past, different types of research papers have been published on molecular docking [6].
From small molecules to large atomic level molecules molecular docking can be used to build a model based on interaction which determines the behaviour, characterization, binding site, and biochemical process of proteins [7].
According to the researcher Meng et al., there are two major steps are involved in molecular docking: first conformation, position, and orientation of ligand can be determined, and second binding affinity of protein-ligand can be estimated. Docking efficiency can be increased if one knows the binding site location in advance. Programs like cavity detection or online servers (GRID, POCKET, SURFNET, PASS, and MMC) can be used to find out about the binding sites of proteins.
In the early days, Fischer developed the lock and key theory for determining the binding sites in ligand - receptors. In this method, the ligand fits with a receptor-like lock and key. Further, Koshland gives the induced fit theory that takes place of the lock and key theory. The induced-fit theory states that the active site of protein is changed its shape according to the interaction of ligand-protein. According to the Koshland theory, ligand-protein interaction should be flexible during molecular docking [1].
The three-dimensional structure of any complex is predicted based on the binding interaction of proteinligand. Various possible poses are generated after the molecular docking is done. The scoring function present in the software grouped and ranked poses together. DNA/protein are used as receptors whereas molecules are used as ligands. The ligand binds specifically at the binding site of the receptor. These interactions are useful in drug design, drug discovery, as well as for mechanistic study. After doing the molecular docking one can tell the information regarding the binding energy, free energy, and stability of the complexes. Databases like protein data banks in employed for searching the suitable protein with accurate PDB format and a methodology to make a PDB file. PubChem is another database that can be used to search different ligands according to the need of our protein.
Tools like DOCK, GOLD, ICM, and FlexX are used for HTS. Based upon the objective of molecular docking the procedure can be either flexible or rigid. Before doing any detailed experimental studies, molecular docking can show the viability of any biochemical reaction. Applications of molecular docking are Lead optimization, Hit identification, and Drug - DNA interaction. There are two major approaches to molecular docking: the simulation approach and the shape complementarity approach. The simulation approach is more flexible and compatible as compared to the shape complementarity approach. But it has one disadvantage also that it takes a longer duration to estimate the docking result [8].
One of the most common brain disorders in aged people is Alzheimer's disease. Alzheimer's disease is a major concern in public health. Alzheimer's disease is undertreated. In 1906 Alois Alzheimer first described this disease. Continuous memory loss is one of the major symptoms. In the beginning, it was thought that Alzheimer’s disease is rare in people but later on it was found that it starts with aging. Memory and language disturbances are seen as clinical symptoms. Other factors include an abnormality in sleep, disturbance in mood, loss of judging ability, etc. Senile plaque and neurofibrillary tangles are the main neuropathological characteristics that are seen in Alzheimer’s disease. As the Alzheimer’s Disease (AD) makes progress the senile plagues that is formed due to the accumulation of beta amyloid is spread to another cortex region. It is observed that in brain first senile plagues are formed. Brain cells are damaged in Alzheimer’s disease. Alzheimer’s disease is thought to as neurodegenerative disease. Neurodegenerative means it initiates the degeneration or it can be defined as loss of neurons in brain. In brain it mainly affects cortex. Tangles and plagues are the two main form that is seen in Alzheimer’s as the disease develop in brain [9].
Alzheimer’s disease is a disorder of neurodegenerative. It affects older people. People over the age of 65 are affected by Alzheimer’s disease. The reasoning ability of people having Alzheimer’s usually decreases as the disease develops. The term that is commonly used for loss in understanding is termed as dementia. Activities of daily life are affected due to the loss of ability. In the United States, Alzheimer’s disease is the sixth major cause of death. As of now, there is no cure for Alzheimer’s disease but there are some treatments available that can enhance some symptoms. Symptoms of Alzheimer's disease depend on the stage of the disease. Alzheimer's disease is classified into preclinical or presymptomatic, mild, and dementia-stage depending on the degree of cognitive impairment. It's become difficult to do motor tasks. Early dementia can be seen in people having trisomy 21. Women have a higher chance of having Alzheimer’s disease after 85 years of age.
In cell membrane of neuron in brain the molecule known as Amyloid Precursor Protein (APP) is present which helps in development and repair of cells. One tail of molecule is present inside the cell and other tail of molecule is present outside the cell. Generally this molecule is get used, break down and gets recycled. The Amyloid Precursor Protein (APP) is get split by two enzymes alpha secretase and gamma secretase. The split up peptide is soluble goes away and everything’s all good. If another enzyme that is beta secretase combine with gamma secretase then this can lead to a serious problem. Now the peptide that is formed is not going to solubilize and it will create a monomer known as amyloid beta. The monomer that is formed aur chemically sticky in nature and they will bond or accumulate outside the neuron to form beta amyloid plagues. This formed plague can get between two or more neuron and can lead to interrupt the neuron signal. If the signals between the brain and body gets interrupt then the person ability will be get affected [10].
In this study, Protein-ligand used for molecular docking is Beta-amyloid and Thioflavin -T. Both in vivo and in vitro Thioflavin-T is used for staining and identifying betaamyloid. Thioflavin-T is a fluorescent dye. The formation of beta-amyloid is seen with the help of fluorescent dye. The binding of Thioflavin-T with beta-amyloid is used as a powerful and easy-to-use tool. Misfolding of amyloid protein is found in Alzheimer’s disease. The protein is insoluble and proteinaceous [11].
In computational drug design and structural molecular biology molecular docking is the main tool. The goal of ligand-protein docking is to predict the predominant binding mode(s) of a ligand with a protein of known threedimensional structure.
Methodology
Autodock Vina
Tools for Autodock Vina:
• Autodock MGL Tools
• Autodock Vina
• PyMOL visualization tool
Steps in Autodock Vina
Choose the protein target and ligand molecule: Here protein that is chosen is Beta-amyloid and the ligand is Thioflavin T for doing molecular docking. The structure of Beta-amyloid protein is downloaded from a database called a Protein Data Bank (PDB) in PDB format. The structure of the Thioflavin T ligand is downloaded from another database called PubChem in SDF format.
Create a folder named docking and place all the dockingrelated files in that folder.
Carry out protein and ligand preparation: Autodock MGL Tool is used for the preparation of protein and ligand.
Protein preparation: Downloaded protein is dragged into the Autodock MGL Tool for the preparation step. Go to edit and delete the entire water molecule. If water molecules are present in the protein pocket region, then the ligand binding is not efficient. Again, go to edit and add polar hydrogen only. The next step is to add charges again and go to edit and add Kollman charges. For saving the protein go to grid click on macromolecule and click on choose and click on the select molecule to save the protein in PDBQT format (Figure 1).
Ligand preparation: First convert the SDF format ligand file into PDB format with the help of the PyMOL visualization tool. Take the ligand file and drag it into the Autodock MGL Tool for the preparation step. Go to ligand click input and click on choose and then select the molecule. The next step is for saving the ligand click on the ligand and go to output to save the ligand in PDBQT format (Figure 2).
Select the docking site in the protein: For preparing the grid drag both the protein and ligand in Autodock MGL Tool. Click on the grid and then select the grid box option. Now the grid box is being created in the center of the protein. Save the grid box file in the grid.txt file.
Carry out protein-ligand docking using a docking tool: Create a config file using a text document. Enter receptor, ligand, center x, center y, center z, size x, size y, size z, energy range, and exhaustiveness in the file. Go to the command prompt for performing the docking. Paste the path of the docking folder which consists of all the files in the command prompt. Now for running the vina paste the path of Autodock Vina in the command prompt. At last for calling out the files from the docking folder type a command (Figure 3).
Different ligand poses are generated: Once the docking is done a log file is generated in the docking folder which consists of 9 poses. These 9 poses tell us about the docking scores. The first pose will be considered the highest affinity score [12,13].
Results and Discussion
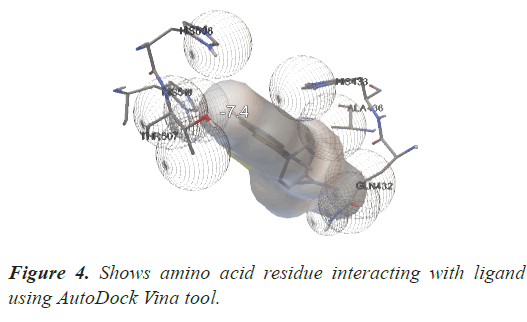
Total 9 best poses of the ligand through which it is bounded with the protein and their energy are generated that tell us about the different molecular scores. The first pose will be considered the highest affinity score. The first pose has energy of – 7.4; the second pose has energy of -7.2, and so on. Ligand–protein will best fit with each other if the binding energy will be low. In this study we have generate an image (Figure 4) which is displaying amino acid residues interacting with ligand using Autodock tool. In analyzing the result of protein and output file in Autodock tool an image is formed which shows amino acid residues THR 507, GLN 432, ALA 436, HIS 510, HIS 433, HIS 506 present at -7.4 docking site (Table 1).
| Protein | Ligand | Docking score | Amino acid residues |
|---|---|---|---|
| Beta amyloid (5tpt) | Thioflavin T (CID-16954) | -7.4 | THR 507, GLN 432, ALA 436, HIS 510, HIS 433, HIS 506 |
Table 1. Analyzing the docking files in AutoDock Vina to find out the amino acid residues present at the highest docking score.
Conclusion
Till today there is no development of drug that is useful in treating Alzheimer's disease. The design of new drug can be beneficial from spreading the beta amyloid protein outside the neuron. In silico tools can be used to design new therapeutic drugs so that the time and cost of the manufacturer industry can be saved. Many different drugs have been rejected or are not successfully run during their clinical trials for treating Alzheimer’s. In this study we have tried to find out the best possible binding site for the interaction of ligand – protein.
References
- Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: A powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 2011; 7: 146-157.
[Crossref] [Google Scholar] [PubMed]
- Jorgensen WL. The many roles of computation in drug discovery. Science 2004; 303: 1813-1818.
[Crossref] [Google Scholar] [PubMed]
- Moitessier N, Englebienne P, Lee D, Lawandi J, Corbeil CR. Towards the development of universal, fast, and highly accurate docking/scoring methods: A long way to go. Br J Pharmacol 2008; 153: S7-S26.
[Crossref] [Google Scholar] [PubMed]
- Bailey D, Brown D. High-throughput chemistry and structure-based design: survival of the smartest. Drug Discov Today 2001; 6: 57-59.
[Crossref] [Google Scholar] [PubMed]
- Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. J Mol Biol 1982; 161: 269-288.
[Crossref] [Google Scholar] [PubMed]
- Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 2004; 3: 935-949.
[Crossref] [Google Scholar] [PubMed]
- McConkey BJ, Sobolev V, Edelman M. The performance of current methods in ligand-protein docking. Current Science 2002; 83: 845-855.
- Dar AM, Mir S. Molecular docking: Approaches, types, applications, and basic challenges. J Anal Bioanal Tech 2017; 8: 356.
- Schachter AS, Davis KL. Alzheimer's disease. Dialogues Clin Neurosci 2000; 2: 91-100.
[Crossref]
- Kumar A, Sidhu J, Goyal A. Alzheimer Disease. StatPearls [Internet] 2022.
- Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta 2010; 1804: 1405-1412.
[Crossref] [Google Scholar] [PubMed]
- Rauf MA, Zubair S, Azhar A. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Int J Basic Appl Sci 2015; 4: 168-177.
- Ayad, Al Dulaimy, Suhad, Hasan. Windows bioinformatics platform: Approach for molecular docking in drug discovery. Int J Sci Engin Res 2015; 6: 5-11.