Research Article - Biomedical Research (2017) Volume 28, Issue 11
Mitochondrial transfer RNA mutations and male infertility
Lin Jin Li1,2, Hang Huang3, Yi Ren Hu2, Hao Chen2, Xiang Chen2, Jun Yang2, Jiaren Li2, Yuan Bo Zheng2, Jian Long Zhu2, Wen Shuo Bao2 and Jun Ouyang1*
1Department of Urology, the First Affiliated Hospital of Soochow University, Suzhou, PR China
2Department of Urology, The Third Clinical Institute Affiliated to Wenzhou Medical University, Wenzhou People’s Hospital, Wenzhou, PR China
3Department of Urology, the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, PR China
- *Corresponding Author:
- Jun Ouyang
Department of Urology
The First Affiliated Hospital of Soochow University, PR China
Accepted date: March 27, 2017
Abstract
Background: Mitochondrial DNA (MtDNA) mutations affecting flagellar movement are often causes of sperm dysmotility. Among these mutations, mitochondrial transfer RNA (mt-tRNA) is the hot spot. However, the role of mt-tRNA mutations in male infertility remains heated debates.
Purpose: To evaluate the roles of mt-tRNA mutations in male infertility.
Methods: To address this problem, we reassessed 5 recent reported mutations: tRNAHis C12187A, tRNAThr G15928A, tRNAArg T10463C, tRNALys A8312G and tRNAAsp T7572C, in clinical expression of asthenozoospermia. We first performed database searches for these mutations; in addition, the phylogenetic approach was performed to determine the Conservation Index (CI) of these mutations between different species. We also used the bioinformatics tool to predict the secondary structure of mttRNAs with and without these mutations. In addition, the pathogenicity scoring system was used to assess the status of each mt-tRNA mutation.
Results: We found that only tRNALys A8312G mutation could be regarded as “possibly pathogenic”, whereas other mutations should be classified as “neutral polymorphisms”.
Conclusions: This is the first report dealing with the association between mt-tRNA mutations and male infertility, thus, our study provided novel insight into the molecular basis underlying mt-tRNA mutations in male infertility.
Keywords
mt-tRNA, Mutations, Male infertility, Pathogenic
Introduction
Infertility affects about 10-15% of all couples attempting pregnancy, and male infertility accounts for half of them. The suspected causes of male infertility include sperm motility disturbances, Y-chromosome microdeletions, chromosomal abnormalities, and hormonal or receptor disorders [1]. Mutation and depletion of mitochondrial DNA (mtDNA) are associated with poor motility and diminished fertility of human sperm [2]. MtDNA is linked to electron transport system and is thus vulnerable to free radical damage and some of its components mutate 100 times more rapidly than nuclear DNA [3].
Human mtDNA is a 16,569 bp double stranded circular DNA molecule coding for 2 rRNAs, 22 tRNAs and 13 polypeptides [4], mtDNA mutations identified in several studies have been associated with poor semen quality, more recently, absence or abnormal mitochondrion has been reported in asthenozoospermic men [5]. Among these mutations, mitochondrial tRNA (mt-tRNA) gene is the hot spot for pathogenic mutations associated with asthenozoospermic men. However, a poor genotype to phenotype correlation is common, as in the case of tRNALeu (UUR) T3291C mutation [6]. Distinguishing the polymorphisms and mutations is important, since fail to do so will inevitably lead to poor diagnosis and genetic advice.
In this study, we reassessed 5 reported mutations: tRNAHis C12187A, tRNAThr G15928A, tRNAArg T10463C, tRNALys A8312G and tRNAAsp T7572C, in clinical expression of asthenozoospermia. We first carried out the database searches for these mutations; moreover, a phylogenetic analysis was used to determine the Conservation Index (CI) of these mutations between different species. We also performed RNA Fold Web Server to predict the Minimum Free Energy (MFE) of these tRNAs with and without these mutations. We finally applied the classic pathogenicity scoring system to discern whether these mutations contributed to the genetic susceptibility of asthenozoospermia.
Materials and Methods
Data extraction
To begin our study, we took the clinical, genetic and molecular datasets from recent publications concerning the association between mt-tRNA mutations and male infertility. Moreover, we searched in the PubMed Central and other public domains with the combination of the following keywords “mitochondrial tRNA mutations, male infertility” or “mt-tRNA variants, male infertility” or “mt-tRNA mutations, asthenozoospermia” or “mt-tRNA variants, oligoasthenozoospermia” to identify the case-control studies examining the link between mt-tRNA mutations and male infertility.
Evolutionary conservation analysis
With the purpose of understanding the molecular basis of mttRNA mutations in male infertility, we performed the phylogenetic analysis for the CI of mt-tRNA mutations. Briefly, 17 vertebrate mtDNA sequences were used for the inter-specific analysis. The CI was then calculated by comparing the human nucleotide variants with other 16 vertebrates. Notably, the CI ≥ 70% was considered as having the functional potential.
Haplogroup classification
As specific mtDNA polymorphisms create groups of related mtDNA haplotypes or haplogroups, to find haplotype/ haplogroup bearing mt-tRNA mutations, we applied the Phylotree to determine the haplotypes/haplogroups [7].
Prediction the MFE structure of mt-tRNA
We further utilized the RNA Fold Web Server program to predict the MFE structure of these mt-tRNAs with and without these mutations (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). In addition to MFE folding, RNA Fold results provided the equilibrium base-pairing probabilities that were calculated via John McCaskill’s partition function algorithm [8,9].
Determining the pathogenicity
McFarland’s team first provided a weighting program for assigning a pathogenicity score to mt-tRNA mutations, their scoring system was revised in 2011 [10,11]. According to that standard, a mt-tRNA mutation will be classified as “neutral polymorphism” if its score is less than 6 points, while the score of a candidate mutation is 7-10, it belongs to “possibly pathogenic”, whereas a mutation with the score more than 11 points, it belongs to “definitely pathogenic”.
Results
Study characteristics
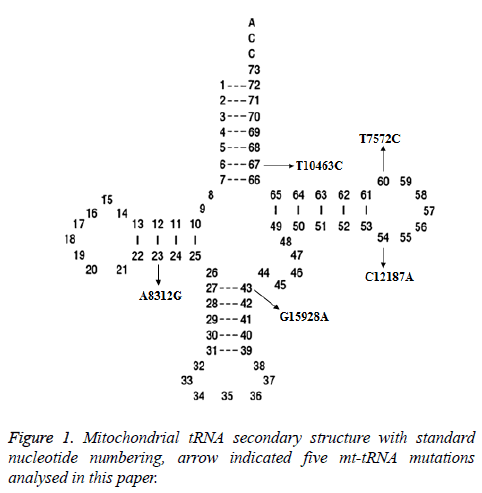
Two potential relevant records were identified by using the keywords mentioned in the previous section. After full-text review, we noticed that one of them was concerning the association between mt-tRNA mutation and asthenozoospermic infertile men from Tunisian population [12]. Whereas another paper which met our inclusion criteria was about the mtDNA polymorphisms in patients with infertility [13]. In total, 5 mt-tRNA mutation have been reported, as shown in Figure 1, the T10463C mutation is localized at the acceptor arm of tRNAArg (position 67), the T7572C and C12187A mutations occur at the T arm of tRNAAsp and tRNAHis genes (position 60 and 54), respectively, the G15928A mutation located at the anticodon stem of the tRNAThr (position 43), while the A8312G mutation is located at the D arm of tRNALys gene (position 23), the characterization of tRNA mutations was listed in Table 1.
| tRNA | Nucleotide changes | Number of nucleotide in tRNA | Position | Homoplasmy or Heteroplasmy | CI (%) | G (wild-type) kcal/mol | G (mutant) kcal/mol | Haplogroup |
|---|---|---|---|---|---|---|---|---|
| tRNAHis | C12187A | 54 | T arm | Homoplasmy | 73 | -11.16 | -11.43 | None-haplogroup specific mutation |
| tRNAThr | G15928A | 43 | Anticodon stem | Homoplasmy | 76.9 | -15.09 | -15.07 | T |
| tRNAArg | T10463C | 67 | Acceptor arm | Homoplasmy | 94.2 | -10.58 | -10.86 | T |
| tRNALys | A8312G | 23 | D arm | Homoplasmy | 96.1 | -16.3 | -18.45 | None-haplogroup specific mutation |
| tRNAAsp | T7572C | 60 | T arm | Homoplasmy | 19.2 | -10.59 | -10.79 | T1 |
Table 1. Molecular characterization of mt-tRNA variants for male infertility.
Conservation assessment of mt-tRNA mutations
Conservation assessment of the bases in the mt-tRNA gene suggests their evolutionary importance; therefore, it is anticipated that mutations may have pathological consequences. In this study, we performed the evolutionary conservation analysis of these mt-tRNA mutations, the CIs of the C12187A, G15928A, T10463C, A8312G and T7572C mutations are 73%, 76.9%, 94.2%, 96.1% and 19.2% respectively. It seems that the T7572C mutation had a very low level of conservation, which ruled out its involvement in the pathogenesis of male infertility.
Haplogroup analysis
By using the Phylotree (http://www.phylotree.org/tree/main.htm), we found that the C12187A and A8312G were nonhaplogroup specific mutations, while both of G15928A and T10463C were the haplogroup T specific variants; moreover, the T7572C belonged to haplogroup T1a1, apparently, the G15928A, T10463C and T7572C were neutral polymorphisms.
Prediction the MFE structure of mt-tRNAs
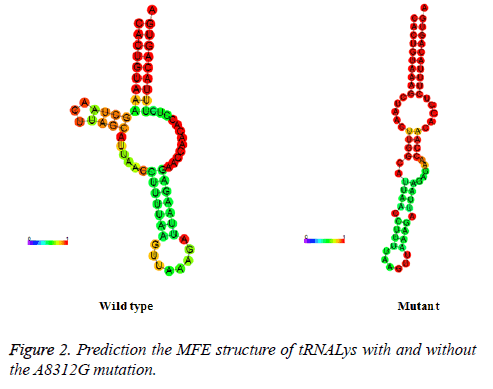
To see whether nucleotide alternations affect the structure of mt-tRNA, the sequences of the wide type and mutant tRNAs harboring the C12187A, G15928A, T10463C, A8312G and T7572C were predicted using the RNA Fold Web Server. As shown in Figure 2, except for the A8312G mutation, other mutations did not affect the secondary structure of the corresponding tRNAs, indicating that the A8312G mutation may play an active role in male infertility.
Pathogenicity scoring system for mt-tRNA mutations
According to the revised pathogenicity scoring system [14], we classified the A8312G mutation as “definitely pathogenic” with a total score of 11 points (Table 2), whereas other mutations, C12187A (3 points), G15928A (5 points), T10463C (4 points) and T7572C (2 points) (data not shown). This indicates that they belong to the “neutral polymorphisms” category.
| Scoring criteria | A8312 mutation | Score/20 | Classification |
|---|---|---|---|
| More than one independent report | Yes | 2 | |
| Evolutionary conservation of the base pair | No changes | 2 | |
| Variant heteroplasmy | Yes | 2 | |
| Segregation of the mutation with disease | Yes | 2 | ≤ 6 points: neutral polymorphisms; |
| Histochemical evidence of mitochondrial disease | No evidence | 0 | 7~10 points: possibly pathogenic; |
| Biochemical defect in complex I, III or IV | No | 0 | ≥ 11 points: definitely pathogenic |
| Evidence of mutation segregation with biochemical defect from single-fiber studies | No | 0 | |
| Mutant mt-tRNA steady-state level or evidence of pathogenicity in trans-mitochondrial cybrid studies | Weak evidence | 3 | |
| Maximum score | 11 | Definitely pathogenic |
Table 2. The pathogenicity scoring system for the A8312G mutation.
Discussion
In mammals, mitochondria are important components of spermatozoa and supply energy for their motility. Indeed, mitochondria are localized in the mid piece of spermatozoa, and deliver ATP, which is mainly used to give flagellar propulsion. To date, more than 150 different mtDNA point mutations as well as a large number of mtDNA deletions are known to be associated with various human mitochondrial diseases [15,16]. However, some of them are single nucleotide polymorphisms and may not play active roles in male infertility, such as the mt-tRNALeu (UUR) T3291C mutation, which is not very conserved from different species, and does not cause the alternation in secondary structure of tRNA Leu (UUR) [6].
For this purpose, we investigated the possible role of mt-tRNA mutations in male infertility. We found that there were 5 mttRNA mutations that had been reported: tRNAHis C12187A, tRNAThr G15928A, tRNAArg T10463C, tRNALys A8312G and tRNAAsp T7572C. Among these, the homoplasmic C12187A and T7572C mutations were localized at the T arm of tRNA genes. While the T10463C and G15928A mutations disrupted the base-pairings (6C-67T, 27A-43G) in the corresponding tRNAs. However, RNA Fold Web Server Program showed that none of them caused the alternation of the secondary structure change of the tRNA genes. On the other hand, the A8312G mutation occurred at the D arm of tRNALys, mt-tRNALys gene was the one of the most mutable genes in mitochondrial genome [17] to date, 16 different pathogenic mutations and 8 polymorphisms in this gene have been identified as causes of various clinical symptoms (http://mamit-tRNA.u-strasbg.fr) [18]. Phylogenetic analysis suggested that the A8312G mutation was not haplogroup specific variant; in addition, this mutation had a very high CI (96.1%). Bioinformatics tool prediction for the secondary structure of tRNALys indicated that it caused a significant change of entropy between the wild type and the mutant, moreover, the pathogenicity scoring system predicted that it had a high score and belonged to a definitely pathogenic mutation, strongly indicated that it may be involved in the pathogenesis of male infertility through the alternation of its secondary structural change, which in turn resulted a failure in tRNA metabolism. Defects in tRNA metabolism will subsequently cause the impairment in mitochondrial protein synthesis.
In summary, this is the first report that reassessing the role of tRNA mutations in male infertility, and our results showed that only the A8312G mutation may be regarded as pathogenic. One of the limitations of the current study was that we did not perform the functional analysis, such as using the cybrid cell containing the A8312G mutation, by examining the steady state level, as well as the aminoacylation ability, to confirm its potential pathogenicity.
Conflict of Interest
None
References
- Yu XW, Wei ZT, Jiang YT, Zhang SL. Y chromosome azoospermia factor region microdeletions and transmission characteristics in azoospermic and severe oligozoospermic patients. Int J Clin Exp Med 2015; 8: 14634-14646.
- Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J Androl 2003; 24: 388-392.
- Chen FF, Zhou CJ, Zhuang CL, Huang DD, Lu JX, Shen X, Chen XL, Yu Z. Mitochondrial energy metabolism disorder and apoptosis: a potential mechanism of postoperative ileus. Int J Clin Exp Med 2015; 8: 14885-14895.
- Aleyasin A, Ghazanfari M, Houshmand M. Leber hereditary optic neuropathy: do folate pathway gene alterations influence the expression of mitochondrial DNA mutation? Iran J Public Health 2010; 39: 53-60.
- Ross A, Christie S, Edmond P. Ultrastructural tail defects in the spermatozoa from two men attending a subfertility clinic. J Reprod Fertil 1973; 32: 243-251.
- Ding Y, Leng J. Is mitochondrial tRNA Leu (UUR) 3291T>C mutation pathogenic? Mitochondrial DNA 2012; 23: 323-326.
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30: 386-394.
- McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 1990; 29: 1105-1119.
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res 2008; 36: 70-74.
- Yarham JW, Al-Dosary M, Blakely EL, Alston CL, Taylor RW. A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat 2011; 32: 1319-1325.
- McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM. Assigning pathogenicity to mitochondrial tRNA mutations: when definitely maybe is not good enough. Trends Genet 2004; 20: 591-596.
- Siwar BG, Myriam G, Afif BM, Emna MR, Nozha C, Afifa S, Faiza F, Leila AK. Two novel mutations in COII and tRNA (His) mitochondrial genes in asthenozoospermic infertiles men. Biochem Biophys Res Commun 2014; 450: 610-615.
- Baklouti-Gargouri S, Ghorbel M, Ben Mahmoud A, Mkaouar-Rebai E, Cherif M. Mitochondrial DNA mutations and polymorphisms in asthenospermic infertile men. Mol Biol Rep 2013; 40: 4705-4712.
- Gonzalez-Vioque E, Bornstein B, Gallardo ME, Fernandez-Moreno MA, Garesse R. The pathogenicity scoring system for mitochondrial tRNA mutations revisited. Mol Genet Genomic Med 2014; 2: 107-114.
- Li C, Lu C, Zhao X, Chen X. Comparison between myocardial infarction and diabetes mellitus damage caused angiogenesis or energy metabolism. Int J Clin Exp Med 2015; 8: 22371-22376.
- Wallace DC. Mitochondrial diseases in man and mouse. Science 1999; 283: 1482-1488.
- Houshmand M, Lindberg C, Moslemi AR, Oldfors A, Holme E. A novel heteroplasmic point mutation in the mitochondrial tRNA (Lys) gene in a sporadic case of mitochondrial encephalomyopathy: de novo mutation and no transmission to the offspring. Hum Mutat 1999; 13: 203-209.
- Putz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA 2007; 13: 1184-1190.