Research Article - Journal of Genetics and Molecular Biology (2021) Volume 5, Issue 2
Mapping of QTLs responsible for yield related traits in advance lines of cotton (Gossypium hirsutum L.).
Hussain SB1, Hussain M1, Sarwar S1, Javed M2* and Zubair M3
1Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Multan, Pakistan
2Department of Plant Breeding and Genetics, Bahauddin Zakariya University, Multan, Pakistan
3Department of Forestry, Bahauddin Zakariya University, Multan, Pakistan
- *Corresponding Author:
- Muhammad Javed
Department of Plant Breeding and Genetics
Bahauddin Zakariya University
Multan, Pakistan
Tel: +92-619210080
E-mail: ch.javedhassan@gmail.com
Accepted on February 21, 2019
Citation: Hussain SB, Hussain M, Sarwar S, et al. Mapping of QTLs responsible for yield related traits in advance lines of cotton (Gossypium hirsutum L.). J Genet Mol Biol. 2019;3(2):11-18.
DOI: 10.35841/genetics-molecular-biology.3.15-22
Visit for more related articles at Journal of Genetics and Molecular BiologyAbstract
Cotton, an important fibre producing crop, enjoys a paramount position among all cash crops. But currently cotton plant is facing genetic erosion due to genetic bottleneck, whereas little effort is carried out in improving cotton genome. Recently many techniques are employed to improve yield to withstand abiotic as well as biotic stresses. Present study was carried out to find loci related to yield traits in cotton genome through association mapping technique. 45 varieties were selected from all over Pakistan and evaluated phenotypically for yield related traits. Correlation between phenotypic traits was calculated. Results of correlation showed that plants having greater number of bolls per plant, number of monopodial and sympodial branches, seed index, boll weight would help breeders to gain more yield. Population structure of all varieties was determined using SSR markers. STRUCTURE software was employed to obtain correct number of sub-populations in graphical form. STRUCTURE analysis divided population, first into 3 main groups and then into 8 groups. Each group showed admixture. General linear modal (GLM) was applied for association mapping. In this approach Q-matrix was used to compensate false positives. QTLs above LOD 2.5 were selected. Five markers gave suitable LOD score. Primer BNL4096 gave highest LOD score of 3.18 for character, yield per plant while primer NAU1070 showed 2.95 LOD score for fibre strength. We found some unique associations of loci with the yield traits of cotton. These associations can further be utilized in increasing the performance of cotton varieties.
Abstract
Cotton, an important fibre producing crop, enjoys a paramount position among all cash crops. But currently cotton plant is facing genetic erosion due to genetic bottleneck, whereas little effort is carried out in improving cotton genome. Recently many techniques are employed to improve yield to withstand abiotic as well as biotic stresses. Present study was carried out to find loci related to yield traits in cotton genome through association mapping technique. 45 varieties were selected from all over Pakistan and evaluated phenotypically for yield related traits. Correlation between phenotypic traits was calculated. Results of correlation showed that plants having greater number of bolls per plant, number of monopodial and sympodial branches, seed index, boll weight would help breeders to gain more yield. Population structure of all varieties was determined using SSR markers. STRUCTURE software was employed to obtain correct number of sub-populations in graphical form. STRUCTURE analysis divided population, first into 3 main groups and then into 8 groups. Each group showed admixture. General linear modal (GLM) was applied for association mapping. In this approach Q-matrix was used to compensate false positives. QTLs above LOD 2.5 were selected. Five markers gave suitable LOD score. Primer BNL4096 gave highest LOD score of 3.18 for character, yield per plant while primer NAU1070 showed 2.95 LOD score for fibre strength. We found some unique associations of loci with the yield traits of cotton. These associations can further be utilized in increasing the performance of cotton varieties.
Keywords
Cotton, Association Mapping (AM), General linear modal (GLM), Population, Structure, Linkage disequilibrium (LD), Quantitative trait locus (QTL).
Introduction
Cotton is currently grown in around 80 countries of the world, covering area about 32-34 million hectares (2011-2012, 2012- 2013). 103.17 million bales of cotton were produced worldwide in 2016 (United States Department of Agriculture 2016). Cotton production is of great importance in Pakistan as it holds 1.0% share in GDP value of Pakistan and bestow 5.1% in whole agriculture value addition. While this year there had been a bad luck towards cotton yield, as it was 27.8% less than the last year production, because of various reasons (Pakistan Bureau of Statistics). It is now crucial to introduce new cotton cultivars that produce enough fibre to meet growing need. Cotton research needs a new and flexile boost to develop high yielding and better-quality cultivars that are more adaptable to the changing environment [1]. Moving from genetic engineering, we came upon a new technique of improving crop plants through marker selection. Markers are special landmarks that show behaviour similar to other landmarks of genome, concerned with expression or phenotype [2]. There are various numbers of markers used for various reasons, like they can differentiate among dominant and co-dominant loci, homozygosity and heterozygosity of particular traits that are under study [3]. Thus we can use markers to identify differences among individuals at the same point by just applying them as they are sensitive to even the slightest change in the genomic sequence also used to find out allele number, gene locations, polymorphism and genetic linkages [4]. Simple sequence repeats (SSR) or microsatellites are more widely used today, these are repetitions in genome ranging from 2 to 5 nucleotides, tandem repeated 5-50 times. Repeats present in the coding regions can give rise to different alleles resulting in change in number of repeats. Advantages of using SSR markers include co-dominance, highly polymorphic among closely related species and present randomly throughout the genome having specific locus. Recent studies showed that these markers are quite reliable for linkage studies [5]. Development of markers from expressed cDNA has proved advantageous as compared to other repeats. EST-SSRs amplifying expressed portion of DNA can be used to find marker trait association more easily [6]. Quantitative traits are those characters that do not show a separate phenotype as compared to the qualitative traits, also the quantitative traits are measured in numbers. The regions expressing these traits are quantitative trait loci or more easily called as QTLs [7]. These regions can vary in number from one to many, controlling a single trait. The study of Quantitative traits is a little difficult, as a proper phenotype is not only the result of the underlying genotype of the individuals, but it also has effect of the surrounding environment under which the individual is grown. To access the exact genotype of the particular plant, it should be grown under controlled environmental conditions [8]. Process of finding order of nucleotides in genome through which sequence and pattern of genes can be identified is called genetic mapping. Mapping is carried out to trace QTLs that cause differences in phenotype. Two methods, linkage mapping and association mapping are very different yet very similar at their roots, because they make use of recombination taking place within genes that ultimately make up a specific phenotype [9].
Association mapping (AM) is a technique based on linkage disequilibrium (LD) analysis, a tool to map those markers that is linked with the traits under study [10]. LD is the correlation between alleles in a mapping population, while linkage is the coinheritance of the loci present near each other. In AM we study the correlation between our selected markers for our trait of interest [11]. This correlation is based on linkage disequilibrium that points out the polymorphisms in the population. Diversity or degree of polymorphism in a population and over all AM study depends upon the strength of LD in the mapping population. The greater the LD the greater the marker and traits are associated [12]. For association analysis LD is measured in natural germplasm, diverse varieties of different origins [13]. Genome wide association mapping involves evaluating the whole genome to test for variations relating to the markers [9]. It requires diverse germplasm and it gives high resolution than linkage mapping because of many recombination’s taking place at many loci at a time [14]. Researchers nowadays are focusing on using association mapping for identifications of QTLs in many plants. First AM analysis was performed on modal plant Arabidopsis thaliana, but now it is executed over various crops like cotton, wheat, rice, maize, potato, barley etc. AM on Arabidopsis was done to find QTLs responsible for its adaptation [15]. In a study 95 diverse species of Arabidopsis was used against 104 markers to identify QTLs related to flowering time [16]. In a study Wheat was used to find out QTLs responsible for its kernel size and milling quality. They used 95 varieties tested for 95 markers to test for QTLs. In another study potato was used to study QTLs responsible for resistance to late blight. They used 123 cultivars and tested them against 49 markers [17]. Association mapping was used to study QTLs responsible for yield components in alfalfa cultivars [18]. Results indicated 15 powerful associations for yield, while a study for sugar beet was carried out on QTLs using GWAS (genome wide association Study) for agronomic trait [19]. Association mapping is a powerful tool for linkage study of QTLs. Our study involves estimation of genotypic diversity among 45 cotton varieties using SSR markers. Analyse association with SSR markers for yield data onto association mapping. The results of this study will help find varieties having QTL’s responsible for higher yield in cotton based on their genotypic framework.
Materials and Methods
In this study 50 seeds from each of these 45 selected varieties were taken from Central Cotton Research Institute Multan. Crop was sown in the agricultural plots of Agriculture Department of BZU. Crop was sown by following Randomized Complete Block Design. Plot size of each variety was kept as 5 lines × 15 feet. All growth factors were taken care of during plant growth.
Trait phenotyping
For phenotyping, 3 of 5 rows were selected and named as R1, R2, R3 (R - replicate) respectively. 3 guarded plants from each row were selected and their phenotypic data was collected. The mean value was calculated further. Yield components evaluated phenotypically include Plant height (PH/cm), 1st Monopodial Node (MN), Number of Monopodial Branches (MB), Number of Sympodial Branches (SB), Number of Bolls (BP), Seed Index (SI, g\100 seeds), Boll Weight (BW/g), Yield per Plant (YP, weight of bolls in g), Ginning Out Turn (G.O.T), Chlorophyll Content (C), Locks per Boll (LB), Seeds per Boll (SB), Uniformity index of lint (UI), Micronaire (M), Strength of the cotton fibre (S) and staple length (SL).
Genotyping
Young leaves from each accession were collected for DNA extraction. Extraction was done as described by [20]. Quantity and quality of extracted DNA was checked on a 0.8% agarose gel before diluting for polymerase chain reaction (PCR) amplification. 41 SSR markers (DPL, JESPR, NAU and BNL) [21,22] of cotton was selected from Cotton Marker Database and were commercially synthesized by Oligo Humanizing Genomics. PCR amplification was performed, and PCR products were separated using 10% non-denaturing polyacrylamide gel electrophoresis. DNA fragments were visualized by staining with silver following procedures described by [20,23].
Statistical analysis
Analysis of Variance (ANOVA) and Correlation analysis of the phenotypic data was done by the Minitab (Table 1). Structure was performed for the evaluation of population structure and kinship analysis. For Structure scoring, presence of band was marked as ‘1’ absence with ‘2’ and missing data was marked ‘99’. 41 SSR markers were used to generate population Structure of the 45 accessions [24], Structure 2.3.1 software was used [25]. Main objective to determine population structure is to avoid false positives during association analysis. Burin length of 10,000 and MCMC run length of 30,000 was used to determine the value of K from a range of 1 to 10. The exact value of the K was determined by running the results on online software the ‘Structure harvester ‘based on estimated log arrhythmic likelihood of natural log of probability data LnP(D). In graph, each value of K shows the estimated K subgroup for mapping population. Structure gives us Q matrix that is further used for TESSEL to carry out GLM to find Marker Trait association. TASSEL 3.0.146 (Trait Analysis by association, Evolution and Linkage) was used to find marker [26]. General Linear Model (GLM) was applied by using Q matrix derived from Structure. Markers that gave significant (LOD=2.5) or highly significant associations (LOD=4) value in general linear model (GLM) was declared as significant associations.
| PH | MN | MP | SP | C | BP | SI | LB | BW | SB | GOT | YP | UI | M | S | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MN | 0.29 | |||||||||||||||
| MP | -0.34 | 0.07 | ||||||||||||||
| SP | 0.31 | 0.06 | 0.09 | |||||||||||||
| C | 0.33 | 0.20 | 0.28 | -0.01 | ||||||||||||
| BP | 0.14 | -0.01 | 0.23 | 0.44 | 0.18 | |||||||||||
| SI | -0.01 | 0.01 | 0.37 | 0.03 | 0.17 | 0.09 | ||||||||||
| LB | 0.23 | -0.03 | -0.17 | 0.11 | 0.35 | 0.11 | -0.02 | |||||||||
| BW | -0.22 | -0.18 | 0.26 | -0.01 | -0.24 | -0.11 | 0.15 | -0.10 | ||||||||
| SB | 0.23 | 0.06 | -0.04 | 0.02 | 0.48 | 0.25 | 0.25 | 0.41 | 0.11 | |||||||
| GOT | 0.28 | 0.03 | -0.08 | -0.22 | 0.28 | -0.18 | -0.13 | 0.18 | 0.14 | 0.01 | ||||||
| YP | -0.19 | -0.14 | 0.20 | 0.29 | 0.04 | 0.50 | 0.26 | 0.06 | 0.32 | 0.15 | -0.22 | |||||
| UI | 0.02 | -0.16 | 0.03 | 0.30 | -0.05 | -0.18 | 0.30 | 0.80 | 0.35 | 0.02 | -0.04 | 0.05 | ||||
| M | -0.08 | -0.27 | -0.13 | -0.22 | 0.00 | 0.24 | -0.04 | -0.02 | -0.08 | -0.11 | 0.07 | -0.13 | -0.14 | |||
| S | -0.11 | 0.10 | 0.16 | -0.24 | -0.06 | -0.13 | 0.03 | 0.16 | 0.30 | -0.01 | 0.01 | -0.08 | 0.02 | -0.08 | ||
| SL | 0.23 | 0.00 | 0.15 | -0.09 | 0.11 | 0.15 | 0.21 | -0.24 | 0.09 | 0.00 | 0.07 | -0.08 | 0.13 | -0.06 | 0.12 | |
Table 1. Correlation analysis of phenotypic traits Plant Height (PH, cm), 1st Monopodial Node (MN), Number of Monopodial Branches (MB), Number of Sympodial Branches (SB), Number of Bolls (BP), Seed Index (SI, g/100 seeds), Boll Weight (BW, g), Yield per Plant (YP, weight of bolls in g), Ginning Out Turn (G.O.T), Chlorophyll Content (C), Locks per Boll (LB), Seeds per Boll (SB), Uniformity index of lint (UI), Micronaire (M), Strength of the cotton fiber (S) and Staple length (SL).
Results
Results include association analysis of 45 cotton genotypes against 41 SSRs markers for cotton. Trait correlation was measured using Minitab 17. Significant positive correlation 0.50 between yield per plant and bolls per plant. This explains that these traits are directly correlated to each other. The lowest negative correlation -0.34 observed was between plant height and monopodials per plant. STRUCTURE 2.3.1software was used to deduce population structure of 45 accessions using 41 SSR markers. Structure software and online software Structure harvester was used to determine graphical representation of sub populations.
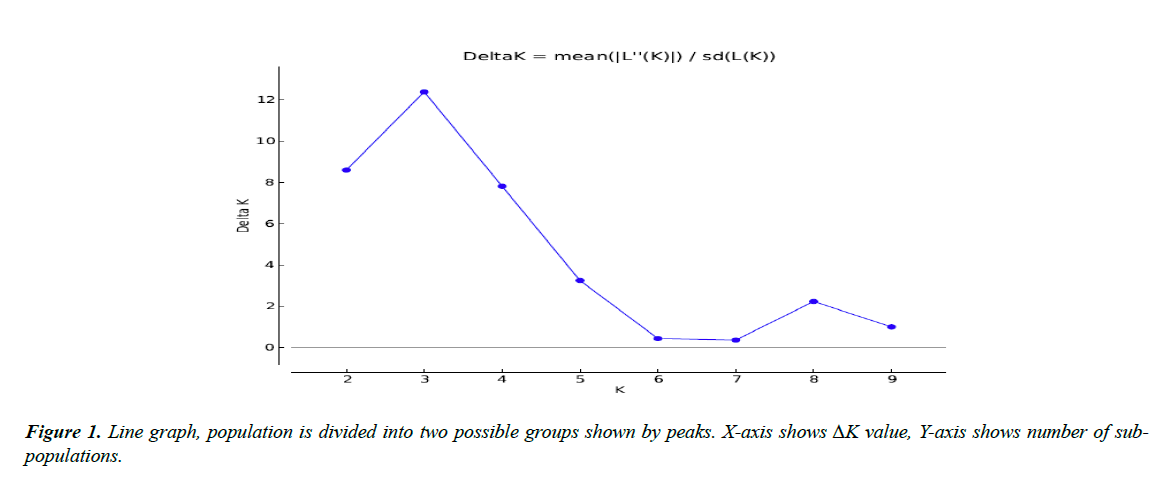
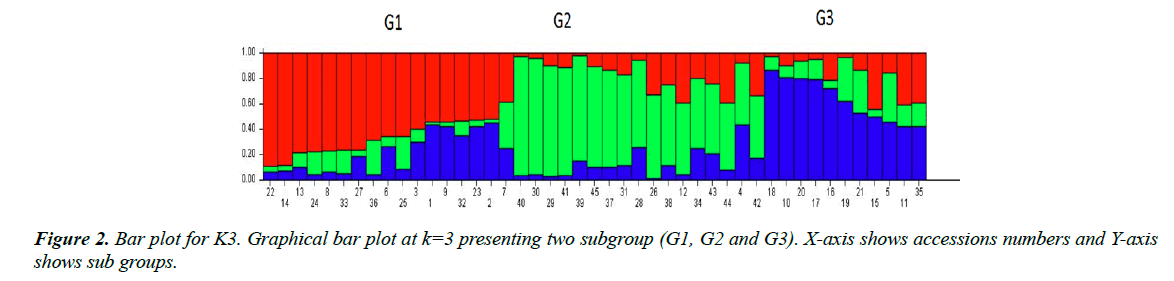
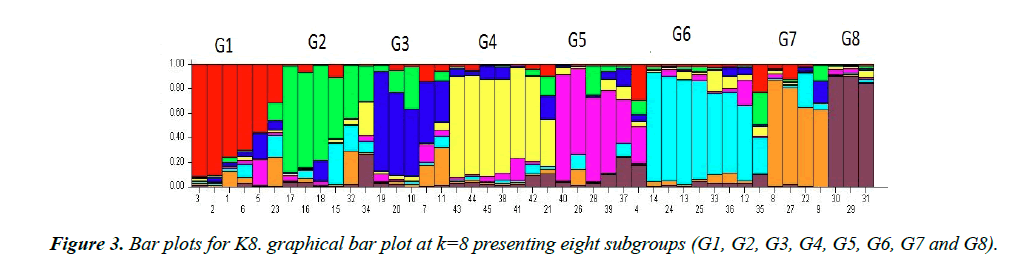
All 45 varieties of cotton were divided into subgroups. Subgroups were determined by the value of LnP(D) “natural log of probability data” using an online software “Structure harvester. Population structure analysis of diverse cotton accessions based on SSR markers is demonstrated through line graph shown in Figure 1. X-axis shows ΔK value, Y-axis shows number of sub-populations. Figure 1 showed two peaks at K=3 and K=8, so we chose both values of ideal grouping of mapping population. Bar plots given below were then taken from STRUCTURE software program by running again at respective peaks.
These bar plots were then divided into subpopulations designated as ‘G’. In K3 as given in Figure 2 showed 3 subgroups each designated as G1, G2 and G3 respectively. G1 subgroup contains 17 varieties. Among them, 13 varieties (CIM-602, CIM-599, CIM-473, CIM-707, CIM-600, CIM-496, N-111, AGC-335, AGC-333, CIM-482, AGC-33, AGC-334 and CIM-446) have their origin from Multan, 3 varieties (FH-142, NIBGE-2 and IR-3701) belong to Faisalabad and 1 variety (CEMB-33) originated from Lahore. G2 subgroup includes 17 varieties. Among them, 10 varieties (SOO-8, CIM-448, CRSM-38, BZU- 75, MNH-886, CIM-598, CIM-622, CIM-506, N-121 and AGC-336) originated from Multan, while 2 varieties (BH-160 and IUB-222) were introduced from Bahawalpur, 1 (CRIS-134) from Sindh, 2 (AA-803, AGS) from Lahore and 2 (FH-14 and NIBGE-3) from Faisalabad. All these varieties were admixed. There were no pure lines. G3 subgroup contained the remaining 11 varieties, among them 7 (CIM-534, CYTO-178, CYTO-177, CIM-616, AGC-337, CIM-573 and CIM-132) originated from Multan, 2 (FH-901 and FH-113) from Faisalabad and 2 (AA- 802 and AA-703) from Lahore. In line graph second peak was observed at K8. Bar plot divided groups into 8 subgroups as showed in Figure 3, designates as G1-G8. G1 group contained 6 varieties, 5 of them (AGC-335, AGC-334, AGC-333, CIM496 and AGC-337) originated from Multan and the remaining one (IR-3701) was from Faisalabad. G2 include 6 varieties. Among them 3 varieties (CIM-616, AGC-33 and CIM-622) were from Multan and 2 varieties (AA-703 and AA-802) were from Lahore and 1 variety (FH-901) was from Faisalabad. G3 group included 5 varieties (CYTO-177, CYTO-178, CIM-534, CIM-446 and CIM-573) originated from Multan. G4 group included 7 varieties, having diverse origin. 3 varieties (CIM- 506, N-121 and MNH-886) were from Multan, 2 (FH-14, FH- 113) from Faisalabad, 1 (AGS) from Lahore and 1 (IUB-222) from Bahawalpur. Sub group G5 includes 6 varieties, only 1 (NIBGE-3) was from Faisalabad while the remaining 5 (SOO- 8, BZU-75, CRSM-38, CIM-448 and AGC-336) were from Multan division. G6 contain 8 varieties, 7 (CIM-602, CIM-559, N-111, CIM-707, CIM-600, CIM-598 and CIM-132) originated from Multan, while the remaining 1 (CEMB-33) was from Lahore. G7 contained 4 varieties of which 2 (CIM-473, CIM- 482) originated from the Multan and the other 2 (NIBGE-2, FH- 142) from Faisalabad. The last group G8 contained 3 varieties, 1 (AA-803) from Lahore, 1 (BH-160) from Bahawalpur and 1 (CHRIS-134) from Sindh. All the varieties groups as K3 and K8 were admixed.
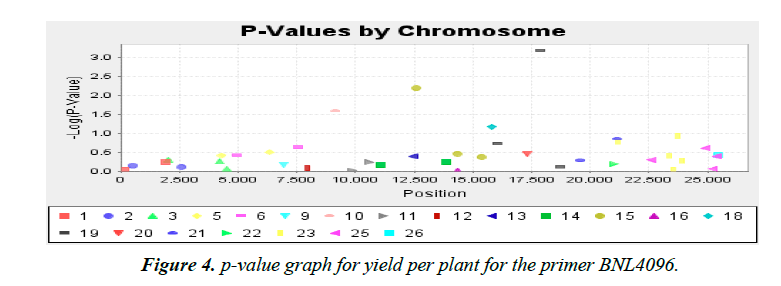
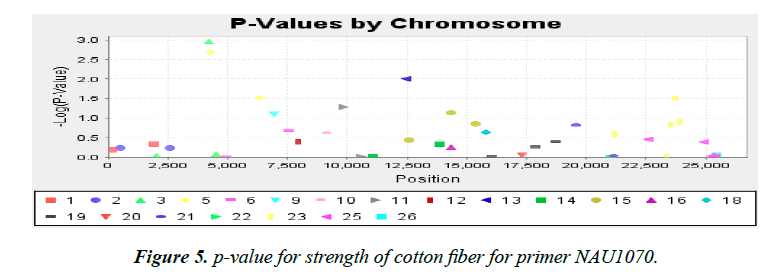
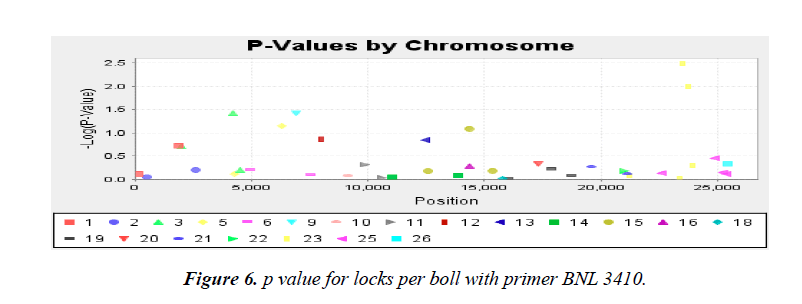
Association analysis was performed with 16 yield traits of cotton using 41 SSR primers. We applied GLM to find association using SSR markers. Q matrix was taken from the population structure analysis to carry out GLM. Number of markers strongly associated with yield components was lower in mapping cotton accessions. P value of the markers gave the idea of associating marker with trait. R2 (phenotypic variance) gave the idea of the degree of QTL effect. Only QTLs having value of LOD above 2.5 were selected for GLM. A little association was observed as only the following traits showed significant LOD value. Markers BNL4096 on chromosomes 19 showed required association with the character yield per plant. The phenotypic variance was 0.17. The observed P value of primer BNL4096 is 6.67 × 10-4 for the respective value of LOD 3.18 for the primer BNL4096 as showed in Figure 4. Marker named NAU1070 at chromosomes 3 showed significant LOD value of 2.94 the phenotypic variance 0.64. The p value of strength of the cotton fibre was 1.1 × 10-3 for the respective value of LOD, 2.69 for primer NAU1070 as showed in Figure 5. Primer BNL 3410 present on chromosome 23 showed LOD values of 2.47 for character Locks per Boll. P value is 3.3 × 10-3. The phenotypic variance showed value of 0.87 as showed in the Figure 6.
Results indicated some useful QTLs related to yield traits of cotton genotypes. Not many QTLs were detected because of small set of SSR markers used. Study shows that association mapping is a powerful tool to detect QTLs in less time that the other laborious methods involved in QTL mapping.
Discussion
Cotton is the most important fibre producing crop, Gaining a prime importance in textile industry. Four species of cotton are cultivated around the world and is very important to improve their genetic base line as these species account for total of 95% of the total cotton production around the world [27,28]. So to improve the genetic makeup of these useful species of cotton various approaches were introduced for increasing yield, quality of the lint, fibre and to increase resistance in the cotton plant to be more resistant towards stresses and diseases [29]. All of these traits mentioned before are quantitative traits so they are controlled by a number of genes present anywhere in the genome. These quantitative traits show continuous variations in their phenotype when observed in segregating populations. The locus that controls the effect of such traits called as QTLs. As present at different positions showing different phenotypic ranges together with environment effect, these QTLs are difficult to be found out [30]. But recent developments in the field of Genetics have paved ways to identify these QTLs with the help of genetic markers.
Newly emerged approach of association mapping has helped a lot in identifying underlying QTLs for many useful traits in plants [31]. Association mapping was previously used in the study of Human Genetics, but now it is successfully employed over plants to unravel the QTLs for major traits in useful and agriculturally important crops [32]. Together with AM, advancements are being made in improving the sequencing techniques for more efficient sequencing of the genomes of the useful crops; all this will ultimately help for GWAS (Genome Wide Association Study), in discovering new and rare alleles and disclosing various epigenetic data about the phenotypic traits of interest [33]. Association mapping or LD mapping will be benefited from this data this can be utilized to find true marker associations, which are linked with the trait of interest [34].
Correlation study of the phenotypic traits of 45 accessions was carried out that showed some significant values of the positive and negative correlation among the traits. In this study significant positive correlation was observed between the traits Yield per Plant and Bolls per Plant. Similar finding was published in the study of [1], showing significant correlation between the two traits. Plant Height also showed some significant correlation with Sympodial branches per Plant and the relative Chlorophyll percentage. Sympodial branches per Plant also showed significant correlation with Bolls per Plant. Chlorophyll content showed correlation with that of Locks per Boll and Seeds per Boll. Seed index showed correlation with Uniformity Index of the cotton fibre.
Population structure was found out using STRUCTURE software for 45 accessions in present study. Population structure was used to define the germplasm in perfect organization, for this STRUCTURE software is being used more frequently. The software involves the genotypic data of the accessions to infer groups in the mapping population using modal based clustering method [30]. According to previous studies various works is done relating to population structure analysis over cotton germplasm around the world [35-38]. Population structure analysis gives us the idea of origin of accessions used for association analysis. In this study total of 41 primers were used against 45 germplasm varieties of cotton to find population structure. The analysis divided the germplasm into 3 groups. All the accessions showed admixture, no variety was pure line. First group contained 17 varieties, of which 13 varieties (CIM-602, CIM-599, CIM-473, CIM-707, CIM-600, CIM-496, N-111, AGC-335, AGC-333, CIM-482, AGC-33, AGC-334 and CIM- 446) originated from Multan, 3 of them (FH-142, NIBGE-2 and IR-3701) belong from Faisalabad and 1 variety (CEMB- 33) originated from Lahore. while the second also contain 17 species of which 10 varieties (SOO-8, CIM-448, CRSM-38, BZU-75, MNH-886, CIM-598, CIM-622, CIM-506, N-121 and AGC-336) originated from Multan, 2 varieties (BH-160, IUB- 222) were introduced from Bahawalpur,1 (CRIS-134) from Sindh, 2 (FH-14, NIBGE-3) from Faisalabad, 2 (AA-803, AGS) from Lahore and the third group contained 11 varieties, among them 7 (CIM-534, CYTO-178, CYTO-177, CIM-616, AGC- 337, CIM-573 and CIM-132) have their origin from Multan, 2 (FH-901 and FH-113) from Faisalabad, 2 (AA-802 and AA- 703) from Lahore. Population structure analysis when carried out at K8, divided the population into 8 subgroups all showing admixed populations.
Association analysis of marker and the required trait is very helpful tool in identifying the QTLs. It depends upon the value of LD between the alleles present at different locus but controlling the same trait [39]. The success of mapping association correlates directly with the value LD between the marker and the alleles governing the trait. The goal of this study was to find associations related to yield and its components in 45 diverse cotton accessions, originated from different areas of Pakistan. We used the GLM based approach to find marker association for yield related traits. GLM requires only the data of population structure to carry out marker trait analysis. We use SSR markers for the association analysis of the cotton accessions related to yield components. Association analysis can be effectively performed over diverse set of germplasm, so it is important to include diverse genotypes in order to get perfect associations with the traits of interest.
Yield per plant in the end defines the total yield of the crop. In our study two markers were found to be associated with the trait yield per plant of the cotton crop. These markers are BNL4096 (p=6.67 × 10-4; R2=0.17) and NAU2437 (p=6.63 × 10-4; R2=0.53). These two markers were detected on chromosome 19 and 15 respectively. In the previous studies various associations were detected using association analysis for yield per plant. These include some significant associations for seed plant yield was found with markers NAU3398-NAU4042, NAU3110- NAU3592, BNL3452-NAU3828 and NAU2700-NAU3588 [1]. A study from china also reported associations with the yield trait with markers reported NAU3269 on chromosome 5, BNL3594 reported on chromosome 25, NAU3100 reported on chromosome 23 and CIR246 reported on chromosome 14 [40].
Strength of cotton fibre is an important factor that accounts for the quality of the cotton grown. In our study two markers NAU1070 (p=11.3 × 10-3; R2 = 0.65) and BNL2662 (p=2.0 × 10-3; R2=0.93). These markers were detected on the chromosomes 3 and 5 respectively in the cotton genome. Study of previous work showed various reported associations of Cotton Fibre Strength with that of SSR markers of cotton. Markers BNL3661, BNL2571, BNL1122 and BNL3806 reported to be associated with that of Strength trait of cotton fibre by [11]. Other studies include some associations reported with markers NAU980, NAU3390 at chromosome 11 by [41]. In another study of association mapping for yield and fibre quality traits they found association of strength of cotton fibre trait with SSR marker given JESPR 153, NAU3736, NAU3778, NAU 5411, BNL827, NAU2894, NAU3110 and NAU3995 [42].
Seeds per Boll give the value of the average seeds produces within each boll. Same is the case only one marker in our study showed significant association with this trait. Marker BNL3171 (p=3.7 × 10-3; R2=0.85) found on chromosome 21 showed significant association value. Previous reported associations include JESPR17 and BNL1059 given by the [43]. In another study they reported marker BNL3259 associated with seeds per boll trait [44].
Phenotypic trait also included the traits locks per boll that gives the average of locks of cotton fibre produced within each boll. Only one marker BNL3410 (p=3.3 × 10-3; R2=0.87) on chromosome 23 was found to be associated with the trait.
Sympodial branches per plant trait was found to be associated with the marker BNL1066 (p=4.2 × 10-3; R2=0.33) present on chromosome 10.
All these results give that association analysis is a tool to find associations with the agronomical important traits among the cash crops. Cotton is among them. Many SSR markers are identified and their associations were found out in the previous studies. These studies are further utilized for making linkage maps and in marker assistant selection studies. The QTLs identified through association mapping can thus be used to improve Cotton varieties with high yielding traits using other techniques like marker assistant selection.
Conclusion
Newly emerged approach, AM has worked a lot in identifying underlying QTLs for many useful traits in plants. Advancements are being made in improving techniques for more efficient sequencing of genomes of useful crops; all of this will ultimately help for GWAS (Genome Wide Association Study), in discovering new and rare alleles and disclosing various epigenetic data about phenotypic traits of interest. Association mapping or LD mapping will be benefited from this data, and can be utilized to find true marker associations, that are linked with the trait of interest.
Association mapping is a tool to find associations with the agronomical important traits of the cash crops. Cotton is one of them. Many SSR markers are identified and their associations were found out in the previous studies. These studies are further utilized for making linkage maps and in marker assistant selection studies. QTLs identified through association mapping can further used to improve Cotton varieties with high yielding traits using other techniques like marker assistant selection
References
- Wang B, Guo W, Zhu X, et al. QTL mapping of yield and yield components for elite hybrid derived-RILs in upland cotton. J Genet Genomics. 2007;34(1):35-45.
- Collard BC, Mackill DJ. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Trans Royal Soc B. 2008;363(1491):557-72.
- Qin H, Chen M, Yi X, et al. Identification of associated SSR markers for yield component and fiber quality traits based on frame map and upland cotton collections. PLoS One. 2015;10(1):e0118073.
- Ferrao LFV, Caixeta ET, Souza FdF, et al. Comparative study of different molecular markers for classifying and establishing genetic relationships in coffea canephora. Plant Sys Evol. 2013;299(1):225-38.
- Han Z, Wang C, Song X, et al. Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet. 2006;112(3):430-9.
- Qureshi SN, Saha S, Kantety RV, et al. EST-SSR: A new class of genetic markers in cotton. J Cott Sci. 2004;8:112-23.
- Ma X, Ding Y, Zhou B, et al. QTL mapping in a-genome diploid asiatic cotton and their congruence analysis with ad-genome tetraploid cotton in genus gossypium. J Genet Genomics. 2008;35(12):751-62.
- Ersoz ES, Yu J, Buckler ES. Applications of linkage disequilibrium and association mapping in crop plants. Genomics assi cro impt. 2007;pp:97-119.
- Paran I, Zamir D. Quantitative traits in plants: Beyond the QTL. Trends Genet. 2003;19(6):303-6.
- Myles S, Peiffer J, Brown PJ, et al. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21(8):2194-202.
- Zhu C, Gore M, Buckler ES, et al. Status and prospects of association mapping in plants. plant genome. 2008;1(1):5-20.
- Abdurakhmonov IY, Saha S, Jenkins JN, et al. Linkage disequilibrium based association mapping of fiber quality traits in G. hirsutum L. variety germplasm. Genetica. 2009;136(3):401-17.
- Chaney L, Sharp AR, Evans CR, et al. Genome mapping in plant comparative genomics. Trends Plant Sci. 2016;21(9):770-80.
- Stich B, Melchinger AE, Frisch M, et al. Linkage disequilibrium in European elite maize germplasm investigated with SSRs. Theor Appl Genet. 2005;111(4):723-30.
- Kump KL, Bradbury PJ, Wisser RJ, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nature Genet. 2011;43(2):163-8.
- Olsen KM, Halldorsdottir SS, Stinchcombe JR, et al. Linkage disequilibrium mapping of arabidopsis CRY2 flowering time alleles. Genet. 2004;167(3):1361-9.
- Tyagi P, Gore MA, Bowman DT, et al. Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L.). Theoretical and applied genetics. 2014;127(2):283-95.
- Malosetti M, Van Der Linden CG, Vosman B, et al. A mixed-model approach to association mapping using pedigree information with an illustration of resistance to phytophthora infestans in potato. Genet. 2007;175(2):879-89.
- Li X, Wei Y, Moore KJ, et al. Association mapping of biomass yield and stem composition in a tetraploid alfalfa breeding population. Plant Genome. 2011;4(1):24-35.
- Zhu Y, Song Q, Hyten D, et al. Single-nucleotide polymorphisms in soybean. Genet. 2003;163(3):1123-34.
- Mei H, Zhu X, Zhang T. Favorable QTL alleles for yield and its components identified by association mapping in Chinese upland cotton cultivars. PLoS One. 2013;8(12):e82193.
- Ullah I, Iram A, Iqbal M.Z, et al. Genetic diversity assessment of cotton (Gossypium hirsutum L.) genotypes from Pakistan using simple sequence repeat markers. Genet Mol Res. 2012;11(1):597-605.
- Shen X, Becelaere VG, Kumar P, et al. QTL mapping for resistance to root-knot nematodes in the M-120 RNR Upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. Theor Appl Genet. 2006;113(8):1539-49.
- Kumar P, Singh R, Lubbers EL, et al. Mapping and validation of fiber strength quantitative trait loci on chromosome 24 in upland cotton. Crop science. 2012;52(3):1115-22.
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14(8):2611-20.
- Pritchard J, Wen X, Falush D. Documentation for structure software (Version 2.3). University of Chicago, Chicago IL. 2010.
- Bradbury PJ, Zhang Z, Kroon DE, et al. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633-5.
- Zhiyuan N, Chen H, Mei H, et al. Molecular tagging of QTLs for fiber quality and yield in the upland cotton cultivar Acala-Prema. Euphytica. 2014;195(1):143-56.
- Gossell-Williams M, Davis A, O'connor N. Inhibition of testosterone-induced hyperplasia of the prostate of Sprague-Dawley rats by pumpkin seed oil. J Med Food. 2006;9(2):284-6.
- Varshney RK, Graner A, Sorrells ME. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005;10(12):621-30.
- Pritchard JK, Stephens M, Rosenberg NA, et al. Association mapping in structured populations. Am J Hum. Genet. 2000;67(1):170-81.
- Wang Q, Fang L, Chen J, et al. Genome-wide mining, characterization, and development of microsatellite markers in gossypium species. Scientific reports. 2015;5(1).
- Taniguchi H, Lowe CE, Cooper JD, et al. Discovery, linkage disequilibrium and association analyses of polymorphisms of the immune complement inhibitor, decay-accelerating factor gene (DAF/CD55) in type 1 diabetes. BMC genetics. 2006;7:22.
- Maskri AY, Sajjad M, Khan SH. Association mapping: A step forward to discovering new alleles for crop improvement. Int J Agric Biol. 2012;15(3):153-60.
- John ZY, Ulloa M, Hoffman SM, et al. Mapping genomic loci for cotton plant architecture, yield components, and fiber properties in an interspecific (Gossypium hirsutum L.× G. barbadense L.) RIL population. Mol Genet Genomic. 2014;289(6):1347-67.
- Tyagi P, Gore MA, Bowman DT, et al. Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L.). Theor Appl Genet. 2014;127(2):283-95.
- Zhao Y, Wang H, Chen W, et al. Genetic diversity and population structure of elite cotton (Gossypium hirsutum L.) germplasm revealed by SSR markers. Plant Syst Evol. 2015;301(1):327-36.
- Fang DD, Jenkins JN, Deng DD, et al. Quantitative trait loci analysis of fiber quality traits using a random-mated recombinant inbred population in Upland cotton (Gossypium hirsutum L.). BMC Genomics. 2014;15(1):397.
- Wilcox PL, Echt CE, Burdon RD. Gene-assisted selection applications of association genetics for forest tree breeding. Asso Map Plants. 2007;pp:211-47.
- Mei H, Zhu X, Guo W, et al. Exploitation of Chinese upland cotton cultivar germplasm resources to mine favorable QTL alleles using association mapping. World cotton germplasm resources. Intech, Rijeka, Croatia. 2014;pp:55-85.
- Zhang T, Qian N, Zhu X, et al. Variations and transmission of QTL alleles for yield and fiber qualities in upland cotton cultivars developed in China. PLoS One. 2013;8(2):e57220.
- Qin H, Chen M, Yi X, et al. Identification of associated SSR markers for yield component and fiber quality traits based on frame map and Upland cotton collections. PLoS One. 2015;10(1):e0118073.
- He DH, Lin ZX, Zhang XL, et al. Mapping QTLs of traits contributing to yield and analysis of genetic effects in tetraploid cotton. Euphytica. 2005;144(1-2):141-9.
- Wang XQ, Yu Y, Li W, et al. Association analysis of yield and fiber quality traits in Gossypium barbadense with SSRs and SRAPs. Genet Mol Res. 2013;12(3):3353-62.