- Biomedical Research (2016) Volume 27, Issue 3
Management of crizotinib resistance in lung cancer using traditional plant source: An in silico strategies.
Anish Kumar, V. Shanthi, K. Ramanathan*Department of Biotechnology, School of Bio Science and Technology, VIT University, India
- *Corresponding Author:
- Ramanathan K
Department of Biotechnology
VIT University
India
E-mail: kramanathan@vit.ac.in
Accepted date: February 29, 2016
Abstract
Crizotinib is an anticancer drug used for the treatment of non-small cell lung cancer (NSCLC). The available evidence suggests that there is development of resistance against crizotinib action due to the emergence of mutations in anaplastic lymphoma kinase (ALK) gene. It is therefore necessary to develop potent anti-cancer drugs for the treatment of crizotinib resistance NSCLC. In the present study, a novel class of lead molecule was screened from plant sources using molecular simulation approach. The bioavailability of the lead compounds was examined with the help of Lipinski rule of five. The toxicity profiles and other physico-chemical properties of drugs were analyzed by OSIRIS program. Additionally, docking strategy was employed to examine the ALK inhibitory activities of the compounds considered in our study. Finally, molecular dynamics simulations were also performed to validate the binding property of the lead compound against native and mutant ALK proteins. Our analysis clearly indicates that CID 9848024 (neoandrographolide) could be the potential ALK inhibitor certainly helpful to overcome the drug resistance in NSCLC. Additionally, the results highlight the importance of traditional plant based compound in the control of drug resistance pattern in cancer.
Keywords
ALK, Crizotinib, NSCLC, Traditional plants, Molecular docking, Molecular dynamics simulation.
Introduction
Lung cancer is the major cause of cancer deaths in the world. Lung cancer is broadly categorized into two main types based upon their histology, which are non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) [1]. The most common forms of NSCLC are adenocarcinoma (ADC) and squamous cell carcinoma (SCC) with NSCLC accounts for about 80% of all lung cancers [2]. The one of the main causes of NSCLC is the chromosomal rearrangements in the anaplastic lymphoma kinase (ALK) gene that codes for anaplastic lymphoma kinase. Mutations in the ALK gene, makes the kinases to send signals to cell to grow and divide in a wild manner. Targeted therapy with ALK inhibitor, crizotinib, may provide great benefits for the treatment of lung cancer. However, prolonged use leads to the development of ALK variant and thus confers crizotinib resistance in patients [3]. According to the study, the echinoderm microtubuleassociated protein-like 4 (EML4)-ALK fusion protein is responsible for lung cancer. An inversion in the chromosome 2 brings together the 5' end of the EML4 gene and the 3' end of the ALK gene resulting in the formation of the EML4-ALK fusion gene. The fusion gene activates fusion proteins and these proteins trigger the signaling pathways which causes lung cancer [4].The experimental evidences indicate that ALK mutations such as C1156Y and L1196M confer a higher level of resistance to crizotinib among the patient [5,6]. These circumstances urge the advancement of new and more successful ALK inhibitors, particularly for the treatment of drug resistant NSCLC [7]. It is believed that plant bio-actives have a great potential to be an inhibitor for the majority of the cases because of its lesser side effects and toxicity [8,9]. In the present analysis, we have screened bioactive compounds from plant sources to overcome the resistance pattern in NSCLC. For instance, we have screened compounds from different plants of Acanthaceae family. The anticancer activity of this plant family is well documented in the recent literature [10]. However, there are no reports available, to highlight the ALK inhibitory activity plants of Acanthaceae family. Therefore, the present study has been aimed to examine the activity of plant bio-actives against ALK. We hope that this approach certainly helpful for the experimental biologists to figure out the potential candidates for NSCLC.
Materials and Methods
Data set
The three-dimensional (3D) structure of native and mutants ALK structures were downloaded from the crystal structures of the Brookhaven Protein Data Bank (PDB) for the analysis [11]. The PDB code for the native structure was 2XP2 whereas for the mutant structures (C1156Y, L1196M, G1269A) PDB codes were 2YJS, 2YFX and 4ANL respectively [12]. The mutant structures (I1171T, S1206Y, G1202R and F1174C) were not available in the Protein data bank, so we have modeled these structures with the help of Swiss PDB viewer. The crizotinib and 22 other plant bioactives from Acanthaceae family were utilized as small molecules in our analysis [13]. The SMILES strings of the crizotinib and the lead compounds were collected from PubChem and submitted to CORINA for developing the 3D structure of molecules [14]. Plants such as Andrographis paniculata, Justicia procumbens, Justicia hyssopifolia, Justicia patentiflora, Rhina canthusnasutus and Hygrophila spinosa would consider for extracting bioactive compounds. The anticancer activities of these plants were reported in the recent literatures [15,16]. However, the activity of these compounds against the treatment of lung cancer is not explored. Hence, bioactives of these plants were utilized for our computational analysis.
ADME and toxicity
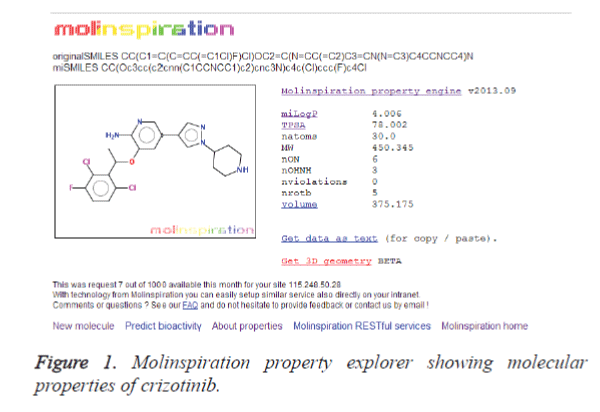
Molinspiration program was used to calculate the molecular properties of the lead compounds in the present study [17]. Toxicity risk assessment of lead compounds is an important parameter should be addressed in the virtual screening strategies. This parameter accounts the majority of the failure of lead compounds in the drug discovery process. Here, we have utilized the OSIRIS program to explore compound toxicity. The OSIRIS program examines the toxicity in terms of mutagenic, tumorigenic and reproductive effects.
Molecular docking
The molecular docking study is vital to understand the bioactivity of the screened lead compounds against the target proteins. In the present study, docking calculation was performed with the assistance of Patch dock algorithm [18]. The energy minimized PDB coordinate file corresponds to the protein and the ligand molecules are the input parameters for the docking analysis. The geometric matching score of crizotinib with target proteins (native and mutant structures) were utilized as a reference for filtering the lead compounds considered in our analysis.
Molecular dynamics simulation
GROMACS Package 4.5.3 implemented with Gromos 43a1 force field was utilized to perform molecular dynamics (MD) of docked complexes such as native-type ALK-crizotinib complex, mutant-type (C1156Y) ALK-crizotinib complex, mutant-type (L1196M) ALK-crizotinib complex, native-type ALK-CID 9848024 complex mutant-type (C1156Y) ALK-CID 9848024 complex and mutant-type (L1196M) ALK-CID 9848024 complex [19,20]. The total simulation time was set to 5,000 ps with integration time step of 2 fs. Structural analysis was done at every picosecond and trajectories were stored in traj.trr file. For instance, root mean square deviation (RMSD) was analyzed with the help of Gromacs utilities g_rms.
Results
Calculation of molecular properties using molinspiration
Molinspiration program was utilized to extract the properties of the lead compounds. The crizotinib properties (Figure 1) were used as a control for screening the other lead compounds. The result is shown in table 1. It is clear from the table that 12 compounds such as CID: 5318517, CID: 11624161, CID: 9848024, CID: 71589914, CID: 11666871, CID: 159982, CID: 460888, CID: 11416971, CID: 5281703, CID: 465430, CID: 5281703 and CID: 5281877 have zero violations for the rule of five. The remaining 10 compounds showed more than one violations for the rule of five. It is bare that for passing oral bioavailability criteria, number of rotatable bonds should be <10 [21]. Therefore, we have made the further refinement of these hits by restricting the number of rotatable bonds to 10. The result is presented in table 2. It is clear from the table 2 that most of the compounds screened from the ADME analysis, possess a reasonable number of rotatable bonds (<10) except CID: 11158514 which showed 12 numbers of rotatable bonds. This result indicates that these 11 compounds may have the potential to become a lead compound for the ALK inhibition.
| S. No. | Compound | miLogP | TPSA | MW | nON | nOHNH | nviolations | volume |
|---|---|---|---|---|---|---|---|---|
| 1 | Crizotinib | 4.006 | 78.002 | 450.345 | 6 | 3 | 0 | 375.175 |
| 2 | CID: 5318517 | 1.050 | 86.99 | 350.450 | 5 | 3 | 0 | 338.33 |
| 3 | CID: 11624161 | 1.72 | 66.76 | 334.46 | 4 | 2 | 0 | 330.29 |
| 4 | CID: 9848024 | 1.17 | 125.69 | 480.60 | 8 | 4 | 0 | 454.37 |
| 5 | CID: 71589914 | 1.75 | 93.07 | 392.49 | 6 | 2 | 0 | 374.84 |
| 6 | CID: 11666871 | 2.87 | 46.53 | 318.46 | 3 | 1 | 0 | 322.24 |
| 7 | CID: 159982 | 3.93 | 72.47 | 394.38 | 7 | 0 | 0 | 333.94 |
| 8 | CID: 460888 | 3.95 | 63.24 | 364.35 | 6 | 0 | 0 | 308.39 |
| 9 | CID: 10458570 | 1.51 | 164.38 | 498.44 | 11 | 5 | 1 | 405.46 |
| 10 | CID: 11416971 | 2.40 | 91.31 | 356.33 | 7 | 1 | 0 | 299.73 |
| 11 | CID: 11785812 | 2.62 | 142.39 | 526.49 | 11 | 3 | 2 | 440.27 |
| 12 | CID: 11478022 | 2.62 | 142.39 | 526.49 | 11 | 3 | 2 | 440.27 |
| 13 | CID: 11284483 | 3.10 | 148.47 | 568.53 | 12 | 2 | 2 | 476.79 |
| 14 | CID: 11158514 | 1.30 | 233.70 | 758.68 | 18 | 4 | 2 | 628.83 |
| 15 | CID: 5281703 | 2.96 | 79.90 | 284.27 | 5 | 2 | 0 | 241.58 |
| 16 | CID: 465430 | 4.33 | 99.14 | 408.41 | 7 | 1 | 0 | 353.14 |
| 17 | CID: 6474554 | 5.79 | 80.67 | 410.51 | 5 | 1 | 1 | 395.92 |
| 18 | CID: 5281703 | 2.96 | 79.90 | 284.27 | 5 | 2 | 0 | 241.58 |
| 19 | CID: 29927693 | 0.15 | 194.96 | 460.39 | 11 | 6 | 2 | 378.60 |
| 20 | CID: 5281877 | 3.97 | 133.52 | 476.57 | 8 | 4 | 0 | 449.89 |
| 21 | CID: 259846 | 8.29 | 20.23 | 426.73 | 1 | 1 | 1 | 461.60 |
| 22 | CID: 5280794 | 7.87 | 20.23 | 412.70 | 1 | 1 | 1 | 450.33 |
| 23 | CID: 72326 | 7.16 | 40.46 | 442.73 | 2 | 2 | 1 | 469.86 |
Bold indicates ADME screened compounds based on Lipinsiki rule of 5
Table 1: Calculations of molecular properties of crizotinib and lead compounds using molinspiration.
| S. No. | Compound | nrotb | S. No | Compound | nrotb |
|---|---|---|---|---|---|
| 1 | Crizotinib | 5 | 13 | CID: 11284483 | 7 |
| 2 | CID: 5318517 | 3 | 14 | CID: 11158514 | 12 |
| 3 | CID: 11624161 | 4 | 15 | CID: 5281703 | 2 |
| 4 | CID: 9848024 | 7 | 16 | CID: 465430 | 7 |
| 5 | CID: 71589914 | 5 | 17 | CID: 6474554 | 9 |
| 6 | CID: 11666871 | 4 | 18 | CID: 5281703 | 2 |
| 7 | CID: 159982 | 4 | 19 | CID: 29927693 | 8 |
| 8 | CID: 460888 | 3 | 20 | CID: 5281877 | 8 |
| 9 | CID: 10458570 | 4 | 21 | CID: 259846 | 1 |
| 10 | CID: 11416971 | 6 | 22 | CID: 5280794 | 5 |
| 11 | CID: 11785812 | 5 | 23 | CID: 72326 | 2 |
| 12 | CID: 11478022 | 5 |
Bold indicates compound with < 10 number of rotatable bonds
Table 2: Details of number of rotatable bonds.
Toxicity prediction using OSIRIS program and molecular docking analysis
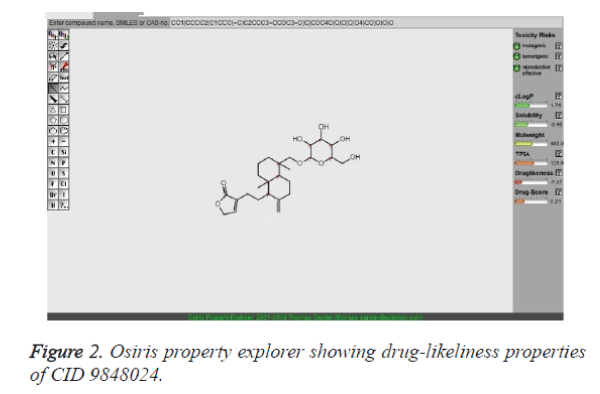
The reason behind the failure of majority of compounds in drug discovery procedure is the issues related to their toxicity. In the present analysis, these issues were addressed with the assistance of the OSIRIS Property Explorer program. The OSIRIS program predicts pharmacokinetics property such as clogP and logS in addition to the toxicity. The result is shown in Table 3. The clogP is a measure of the compound's hydrophilicity. The high clogP qualities may bring about poor retention due to the compound's low hydrophilicity. The data assessed in table 3 demonstrates the mutagenic, tumorigenic and reproductive effects of lead compounds. It is clear from the table that compounds such as CID: 5318517, CID: 11624161, CID: 9848024, CID: 71589914 and CID: 11666871 did not show mutagenic, tumorigenic and reproductive effect when run through the OSIRIS program.
| S.No. | COMPOUND | Mutagenic | Tumorigenic | Reproductive Effective | cLogP | Solubility | Drug Likeness | Drug Score |
|---|---|---|---|---|---|---|---|---|
| ID | ||||||||
| 1 | Crizotinib | No | No | No | 3.54 | -5.26 | 3.12 | 0.52 |
| 2 | CID: 5318517 | No | No | No | 1.88 | -2.95 | -4.59 | 0.43 |
| 3 | CID:11624161 | No | No | No | 2.72 | -3.35 | -4.83 | 0.25 |
| 4 | CID: 9848024 | No | No | No | 1.74 | -3.45 | -7.37 | 0.21 |
| 5 | CID:71589914 | No | No | No | 2.37 | -3.36 | -4.37 | 0.4 |
| 6 | CID: 11666871 | No | No | No | 3.58 | -3.75 | -10.93 | 0.23 |
| 7 | CID: 159982 | No | Yes | No | 3.47 | -6.45 | 0.56 | 0.24 |
| 8 | CID: 460888 | No | Yes | No | 3.54 | -6.43 | 0.6 | 0.24 |
| 9 | CID: 11416971 | Yes | No | No | 2.51 | -5.23 | -2 | 0.22 |
| 10 | CID: 5281703 | Yes | No | No | 2.61 | -3.17 | 0.87 | 0.44 |
| 11 | CID: 465430 | No | No | Yes | 4.26 | -4.99 | -6.25 | 0.1 |
| 12 | CID: 5281703 | Yes | No | No | 2.61 | -3.17 | 0.87 | 0.44 |
Table 3: Toxicity risks and physicochemical properties of crizotinib and lead compounds predicted by OSIRIS Property Explorer.
Therefore, these compounds were further validated by means of molecular simulation study. The lead compounds such as CID: 5318517, CID: 1162416, CID: 9848024, CID: 71589914 and CID: 11666871 were used for docking analysis. Docking analysis was performed twice to eliminate the false positive. The result is shown in table 4.
| S. No. | Compound | Native (2XP2) | L1196M (2YJS) | C1156Y (2YFX) | G1269A (4ANL) | I1171T* | S1206Y* | G1202R* | F1174C* |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Crizotinib | 5042 | 4728 | 4928 | 5128 | 4850 | 4938 | 5016 | 4850 |
| 2 | CID: 5318517 | 4114 | 4188 | 4364 | 4360 | 4070 | 4032 | 4138 | 4314 |
| 3 | CID:11624161 | 4034 | 4194 | 4160 | 4584 | 4060 | 4126 | 4080 | 4034 |
| 4 | CID:9848024 | 5342 | 5046 | 5912 | 5442 | 5462 | 5568 | 6044 | 5362 |
| 5 | CID: 71589914 | 4676 | 4470 | 4844 | 4922 | 4778 | 4778 | 4778 | 4778 |
| 6 | CID:11666871 | 4008 | 4046 | 4210 | 4256 | 3882 | 4266 | 4266 | 4042 |
* indicates modeled structure with the help of Swiss PDB viewer.
Table 4: Docking score of the crizotinib and lead compounds against the target structure.
The docking score of native-type ALK-crizotinib complex was found to be 5042 and for the mutant types ALK (C1156Y and L1196M)-crizotinib complex docking score were found to be 4728 and 4928 respectively. The docking score of G1202Rcrizotinib complex was found to be 5016 and for G1202R-CID 9848024 was found to be 6044. The docking score of F1174Ccrizotinib complex was found to be 4850 and for F1174C-CID 9848024 was found to be 5362.
It is clear from the results that CID: 9848024 (neoandrographolide) from our dataset showed higher docking score not only against native type ALK but also with mutant types ALK studied. For instance, the docking score of nativetype ALK-CID: 9848024 complex was found to be 5342 and for the mutant types ALK (C1156Y and L1196M)-crizotinib complexes score were found to be 5046 and 5330 respectively.
Further validation of CID 9848024 compound was done with the help of molecular dynamics simulation study.
Molecular simulation analysis
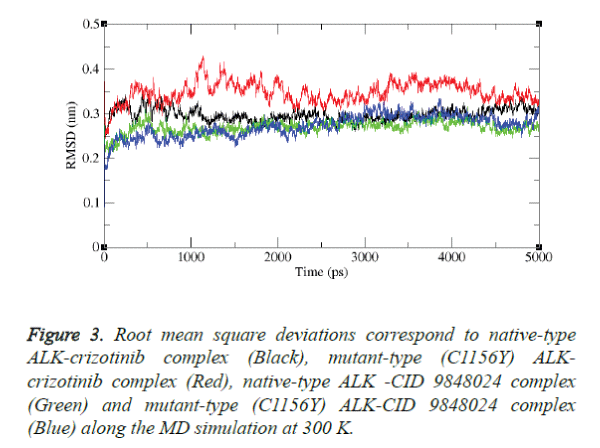
Molecular dynamics simulation study was carried out with the aid of GROMACS package 4.5.3 to explore the stability of the complex structures over the simulation time. In particular RMSD was examined from the trajectory file. The results were shown in figures 3 and 4.
The results depict that native type ALK-crizotinib complex structure acquired ~0.30 nm, while mutant type (C1156Y) ALK-crizotinib complex structure acquired ~0.38 nm of backbone RMSD at 1000 ps (Figure 3).
On the other hand, native-type ALK-CID 9848024 structure acquired ~0.25 nm of backbone RMSD while mutant-type (C1156Y) ALK-CID 9848024 complex structure acquired ~0.23 nm of backbone RMSD at 1000 ps. Between a period of 2000 and 4000 ps, native type ALK-crizotinib complex structure maintains a RMSD value of ~0.29 nm whereas mutant type (C1156Y) ALK-crizotinib complex structure showed a deviation from ~0.36 nm to ~0.38 nm.
On the other hand, native-type ALK-CID 9848024 structure maintains a RMSD value of ~0.24 nm and mutant type (C1156Y) ALK-CID 9848024 complex structure showed a RMSD value between ~0.24 nm to ~0.27 nm.
Finally, mutant type (C1156Y) ALK-crizotinib complex structure attained RMSD of ~0.31nm and native type ALKcrizotinib complex structure attained RMSD of ~0.27 nm whereas native-type ALK-CID 9848024 complex structure attained RMSD of ~0.24 nm and mutant-type (C1156Y) ALKCID 9848024 complex structure attained RMSD of ~0.29 nm at the end of the simulation period.
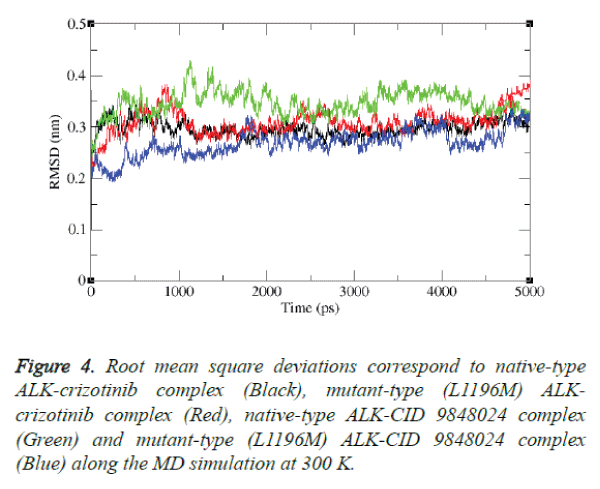
The stability of L1196M-neoandrographolide complex was also examined by simulation study. The result is shown in figure 4.
It is evident from the figure that after the relaxation period native type ALK-crizotinib complex structure acquired ~0.30 nm, while mutant-type (L1196M) ALK-CID 9848024 complex structure acquired ~0.32 nm of backbone RMSD at 1000 ps.
It is noteworthy to mention that native type ALK-CID 9848024 complex structure attained the RMSD value of ~0.34 nm while mutant-type (L1196M) ALK-CID 9848024 complex structure attained the RMSD value of ~0.25 nm at 1000 ps. Between 2000 and 4000 ps, native type ALK-crizotinib complex structure maintains a RMSD value of ~0.28 nm whereas mutant type (L1196M) ALK-crizotinib complex structure maintains a RMSD value of ~0.30 nm. On the other hand, native-type ALK-CID 9848024 structure showed a RMSD value between ~0.36 nm to ~0.38 nm and mutant type (L1196M) ALK-CID 9848024 complex structure showed a RMSD value between ~0.24 nm to ~0.26 nm between 2000 and 4000 ps.
At the end of 5000 ps, the mutant type (L1196M) ALKcrizotinib complex structure attained RMSD of ~0.37 nm and native type ALK-crizotinib complex structure attained RMSD of ~0.26 nm whereas native-type ALK-CID 9848024 complex structure attained RMSD of ~0.33 nm and mutant-type (L1196M) ALK-CID 9848024 complex structure attained RMSD of ~0.32 nm.
Discussion
Drug resistance in crizotinib is a global issue needs to be addressed. To overcome drug resistance in NSCLC, virtual screening studies are applied to identify some novel compounds for drug resistance lung cancer types. The initial screening was accomplished by means of Lipinski rule of five. The rule states that most molecules with good membrane permeability should have a molecular weight ≤ 500, calculated octanol–water partition coefficient, log P ≤ 5, hydrogen bond donors ≤ 5, acceptors ≤ 10 and van der Waals bumps polar surface area (PSA) <120 Å2 [22]. This brings to the conclusion that the bioavailability of former 12 compounds was significantly better than the later 10 compounds in our dataset.
It has been exhibited that for compounds to have a probable reason of being well absorbed, their clogP value should not be more than 5.0. It is clear from the table 3 that clogP values of the 11 compounds found to be in the acceptable criteria. Of note, solubility of the lead compounds was also found in the comparable zone (less than -4.0) with that of standard drugs to fulfill the requirements. Subsequently, the molecular docking program was employed to find out the binding affinity of lead compounds with the target proteins. The lesser docking score implies that mutation results loss of binding affinity for crizotinib. It is believed that a potential lead compound is the one should have higher docking scoring than the existing drug molecule, crizotinib. Therefore, we have examined docking score for all the hits both with the native type and with mutant type ALK systems. This result highlights that CID 9848024 has a better binding affinity with both native type and mutant ALK system than crizotinib. In essence, the pharmacokinetics and pharmacodynamic properties of CID 9848024 results better than the other lead compounds considered in our study (Table 3). The drug-likeliness property of CID 9848024 is shown in Figure 2 with the help of Osiris property explorer. It is noteworthy that our lead compound CID 9848024 (neoandrographolide) is a compound isolated from the Andrographis paniculata plant which could possess anti lung cancer activity. Finally, RMSD investigation provides an understanding of how much the three-dimensional structure fluctuates over the simulation time. Overall, the lesser RMSD values of ALK- neoandrographolide complexes than ALK - crizotinib complexes clearly highlights the potential of neoandrographolide in the inhibition of ALK and its variants. `
Conclusion
In the present investigation, we have examined the activity of neoandrographolide against native and mutant type ALK proteins by employing computational strategies. The result from our analysis highlights that neoandrographolide showed zero violations to the rule of five. Moreover, it was found that neoandrographolide did not show toxicity when run through the OSIRIS program. Additionally, docking study showed that neoandrographolide has the highest binding affinity not only with native type ALK but also with the mutant type ALK systems (C1156Y and L1196M) among the lead compounds considered in our study. Finally, RMSD data acquired from molecular dynamic simulation concludes the stable binding of neoandrographolide with ALK proteins. Overall, the results from our study clearly indicate that neoandrographolide could be certainly helpful to overcome the Lung cancer drug resistance in the near future. However, caution is required in such interpretation and experimental study will be necessary to confirm the conclusions.
Acknowledgements
The authors express a deep sense of gratitude to the Management of Vellore Institute of Technology for all the support, assistance and constant encouragement provided by them to carry out this work.
References
- Siegel R, Naishadham D, Jemal A. Cancer Statistics. Cancer J Clin 2012; 62: 10-29.
- Skarda J, Marian H, Vítezslav K. Drug resistance in lung cancer. Cancer Ther 2008; 6: 377-388.
- Sun H, Li Y, Li D, Hou T. Insight into crizotinib resistance mechanisms caused by three mutations in ALK tyrosine kinase using free energy calculation approaches. J ChemInf Model 2013; 53: 2376–2389.
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALKfusion gene in non-small-cell lung cancer.Nature 2007; 448: 561-566.
- Katayama R, Alice TS, Tahsin MK, Mari MK, Benjamin JS, Balazs H, Nicholas AJ, John CW, Alan TY, Cyril B, Lisa D, Jamal Carlos S, Katherine C, Lecia VS, John I, Jeffrey AE. Mechanisms of acquired crizotinib resistance in ALK rearranged lung cancers. SciTransl Med 2012; 4: 120.
- Kumar A, Shanthi V, Ramanathan K. Computational investigation and experimental validation of crizotinib resistance conferred by C1156Y mutant anaplastic lymphoma kinase. MolInf 2015; 34: 105-114.
- Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer.Clin Cancer Res 2011; 17: 2081-2086.
- Saadat F, Sardari S, Maleki B. Virtual screening of anti-mycobacterial plant compounds. MolInf 2013; 32: 802-810.
- Niranjan A, Tewari SK, Lehri A. Biological activities of Kalmegh (AndrographispaniculataNees) and its active principles-A review. Indian J Nat Prod Resour 2010; 1: 125-135.
- Awan JA, Ahmed BC, Uzair M, Aslam SM, Farooq U, Ishfaq K. Family acanthaceae and genus aphelandra: ethnopharmacological and phytochemical review. Int J Pharm PharmSci 2014; 6: 10.
- Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. The protein data bank. Nucleic Acids Res 2000; 28: 235-242.
- Cui JJ, Tran-Dube M, Shen H, Nambu M, Kung PP, Pairish M, Jia L, Meng J, Funk L, Botrous I, McTigue M, Grodsky N, Ryan K, Padrique E, Alton G, Timofeevski S, Yamazaki S, Li Q, Zou H, Christensen J, Mroczkowski B, Bender S, Kania RS, Edwards MP. Structure based drug design of crizotinib, a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem 2011; 54: 6342-6363.
- Feldman J, Snyder K A, Ticoll A, Pintilie G, Hogue CW. A complete small molecule dataset from the protein data bank. FEBS Lett 2006; 580: 1649-1653.
- Gasteiger J, Rudolph C, Sadowski J. Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Comput Method 1990; 3: 537-547.
- Chao WW, Lin FB. Isolation and identification of bioactive compounds in Andrographispaniculata (Chuanxinlian). Chinese Medicine 2010; 5: 17.
- Kshirsagar DA, Ingale GK, Vyawahare SN, Thorve SV. Hygrophilaspinosa: A comprehensive review. Pharmacogn Rev 2010; 4: 167-171.
- Buntrock RE. ChemOffice Ultra 7.0. J ChemInfComputSci 2002; 42: 1505-1506.
- Schneidman D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 2005; 33: 363-367.
- Hess B, Kutzner C, Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput2008; 4: 435-447.
- Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen H J, GROMACS: fast, flexible, and free. ComputChem 2005; 26: 1701-1718.
- Oprea TI. Property distribution of drug-related chemical databases. J Comput Aided Mol Des 2000; 14: 64-251.
- Muegge I. Selection criteria for drug-like compounds. Med Res Rev 2003; 23: 302-321.