- Biomedical Research (2014) Volume 25, Issue 1
?-lactamase gene blaSHV detected in bacteria isolated from retail sushi in Kampar, Malaysia.
Hui-Ting Cheong1, Wai-Yew Ho2, Quok-Cheong Choo3, and Choy-Hoong Chew1*
1Department of Biomedical Science, Faculty of Science, Universiti Tunku Abdul Rahman, Jalan Universiti, Bandar Barat, 31900 Kampar, Perak, Malaysia.
2Department of Science and Engineering, Centre for Foundation Studies, Universiti Tunku Abdul Rahman, Jalan Universiti, Bandar Barat, 31900 Kampar, Perak, Malaysia.
3Department of Biological Science, Faculty of Science, Universiti Tunku Abdul Rahman, Jalan Universiti, Bandar Barat, 31900 Kampar, Perak, Malaysia.
- *Corresponding Author:
- Choy-Hoong Chew
Department of Biomedical Science,
Faculty of Science,
Universiti Tunku Abdul Rahman
Jalan Universiti, Bandar Barat,
31900 Kampar, Perak, Malaysia
Accepted date: September 17 2013
Citation: Hui-Ting Cheong, Wai-Yew Ho, Quok-Cheong Choo and Choy-Hoong Chew. β-lactamase gene blaSHV detected in bacteria isolated from retail sushi in Kampar, Malaysia. Biomedical Research 2014; 25 (1): 25-31.
Abstract
Increasing prevalence of multi-antibiotic resistant bacteria in humans is a major concern for public health. The widespread use of antimicrobial agents in plant production, animals and food production has created a reservoir of resistant bacteria with resistance genes that can be transmitted to humans through foodborne route. Hence, the priority of this study was to isolate and characterize multi-antibiotic resistant bacteria from retail sushi in Kampar. Twenty sushi samples consisting of egg, shrimp, cucumber, raw salmon, and fish roe were sampled. A total of 22 bacteria colonies were isolated on nutrient agar supplemented with ampicillin. Gram staining, biochemical tests and Kirby-Bauer antibiotic susceptibility tests were carried out to investigate characteristics of bacterial isolates. From the 22 bacterial isolates, 45% (10/22) were found to be multi-antibiotic resistant. All bacterial isolates were then cultured overnight with ampicillin-supplemented nutrient broth and total DNA was extracted from each using the rapid boil method. The extracted DNA was used as a template for PCR amplification of β-lactamase gene using blaSHV and blaTEM specific primers. Surprisingly, 9% (2/22) of the multi-antibiotic isolates were tested positive for blaSHV gene, while none carried blaTEM gene. The bacteria harboring blaSHV was identified to be Raoultella ornithinolytica through API 20E test. The presence of β-lactamase gene blaSHV in the bacteria isolates from sushi may be a source of transmission of antibiotic resistance traits in human via the food chain.
Keywords
β-lactamase, blaSHV, blaTEM, sushi, foodborne, antibiotic resistance
Introduction
Large amounts of antibiotics are used in human therapies, farm animals and aquaculture as growth-enhancing compounds. Antibiotics are also used increasingly in plant biotechnology for the treatment of bacterial diseases [1-2]. As a result, bacteria are constantly subjected to selection pressure and acquire resistance to multiple drugs. The resistant bacteria may be transmitted to humans and cause diseases that cannot be treated using conventional antibiotics [3].
One of the first reports of resistance in food was reported by Starr and Reynolds [4-5]. It was shown that turkeys fed with streptomycin-containing diet carried bacteria that were resistant to streptomycin antibiotic compared to the turkeys fed with non-antibiotic-supplemented feed. As food from animals is considered a major reservoir of antibiotic-resistant bacteria, researchers are continuously monitoring the patterns of resistance in food from animals. For instance, methicillin-resistant Staphylococcus aureus was isolated in 31/402 (7.7%) of retail pork samples from four Canadian provinces [6]. In Vietnam, a study demonstrated that 50.5% of Salmonella spp. and 83.8% of Escherichia coli from raw food were resistant to at least one antibiotic [7]. The outbreak of Shigella sonnei infection in Norway, Sweden, and the United Kingdom in 1994 incriminated imported iceberg lettuce as the vehicle of transmission[8]. Studies have also documented the emergence of nalidixic acid resistant Shigella dysenteriae type 1 in coconut milk dessert [9].
Specifically, β-lactamases represent the most common cause of bacterial resistance to β-lactam antibiotics because these antibiotics are the preferred treatment regimens for many infectious diseases [10-11]. TEM and SHV are both examples of class A β-lactamases, the earliest class of β-lactamase which was discovered and the most prevalent class among existing pathogens today [12]. TEM- and SHV-type β-lactamases have been detected in clinical isolates around the world [13-17]. Molecular analysis of antibiotic resistance genes and antibioticresistant mobile elements showed that identical elements could be found in bacteria that colonize both animals and humans [7]. This provides clear evidence of a transfer of these resistance genes between human and animal bacteria. As sushi is mostly made out of raw materials, it was hypothesized that it could be a reservoir for resistance genes dissemination to humans via the food chain. This study here reports the isolation of multi-antibiotic resistant bacteria, and detection of the β-lactamases gene blaSHV in bacteria isolated from ready-to-eat sushi in Kampar, Malaysia.
Material and Methods
Sushi Samples and Bacterial Isolation
Different kinds of sushi were selected and targeted for this study: cucumber, egg, fish roe, raw salmon and shrimp sushi. All samples were collected from a hypermarket and a Japanese restaurant in Kampar, Perak Malaysia. Direct inoculation method was used in which the surface of sushi samples were swabbed thoroughly with sterile cotton swab before streaking onto ampicillin (Sigma Aldrich, USA)-supplemented nutrient agar (Merck, USA). Agar plates were then incubated overnight in 37ºC incubator. The following day, single colonies with different morphological characteristics were selected and propagated onto fresh agar plates supplemented with ampicillin.
Gram Staining and Biochemical Tests
A single colony from a pure culture was mixed with one drop of sterile saline and smeared on a microscope slide for Gram stain (LabChem, Malaysia). All bacterial strains isolated were also tested using a range of general biochemical tests such as motility, hydrogen sulfide test, oxidation- fermentation (OF), oxidase and catalase tests. API 20E Bacteria Identification Kit (bioMerieux®, USA) was utilized to identify the bacteria isolates.
Kirby-Bauer disk diffusion susceptibility test
From each pure culture, the turbidity of the inoculum was adjusted to 0.5 McFarland standard and swabbed on Muller- Hinton agar using a sterile swab. For each plate, six types of antibiotic discs for ampicillin (Am), amikacin (Ak), ceftriaxone (CRO), chloramphenicol (C), tetracycline (TE) and trimethoprim (W5) (BD BBL™ Oxoid, UK) were applied onto the surface of agar using sterilized forceps. The inoculated plates were incubated agar side up in a 37°C incubator for 16-18 hours. The zone of inhibition was measured after incubation for 16 to 18 hours in 37°C incubator according to the Clinical and Laboratory Standards Institute (CLSI) standard (Performance Standards for Antimicrobial Susceptibility Testing; Twenty- First Informational Supplement, 2011).
Total DNA Extraction and Polymerase Chain Reaction (PCR)
Total DNA was extracted using modified fast boil method in accordance to Kor et al. (2013) [18]. The integrity of bacterial DNA was tested using 16S rRNA primer [19]. Using gene-specific forward and reverse primers of blaTEM and blaSHV, PCR was carried out to detect these antibiotic resistance-conferring genes. The primer sequences and expected sizes of amplicons were obtained from other studies [20-21]. A positive and negative standard for the respective primers were included in each PCR, using cycle conditions of 94ºC for 180 s, followed by 35 cycles of 94ºC for 45 s, 55ºC for 60 s, 72ºC for 50 s, and the final elongation 72ºC for 300 s (for 16S rRNA amplification) or 94ºC for 300 s, followed by 30 cycles of 94ºC for 45 s, 55ºC for 45 s, 72ºC for 45 s, and the final elongation 72ºC for 300 s (for blaTEM amplification). For blaSHV amplification, the cycle conditions were similar to blaTEM, except the annealing temperature was 50ºC. The concentration of reagents in PCR reaction mixture was 1X PCR Buffer, 2 mM of MgCl2 (Invitrogen, USA), 0.4 mM of each forward and reverse primers, 0.2 mM of dNTP mix (Promega, USA), 0.04 U/μl of Taq polymerase (Invitrogen, USA) and 15 ng/μl of DNA template in a final volume of 25 μl. The final products were electrophoresed on 1.5% agarose gel, stained with EtBr before visualization in a UV transilluminator (Syngene, UK).
Results
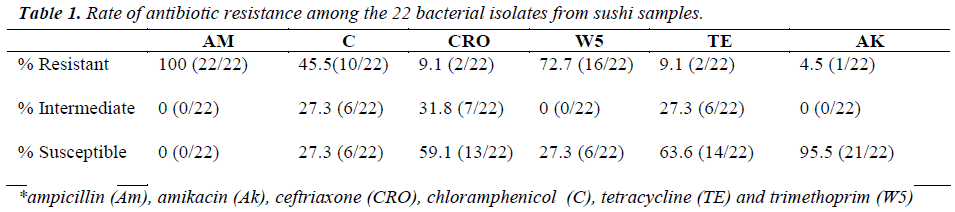
In this study, the isolates were either coccobacillus or bacillus gram-negative bacteria, except for one isolate. The diameter for zone of inhibition to the nearest millimeter was measured for each antibiotic tested and summarized in Table 1. From this study, all of the isolates (100%)
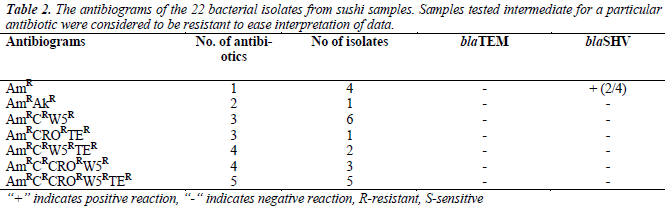
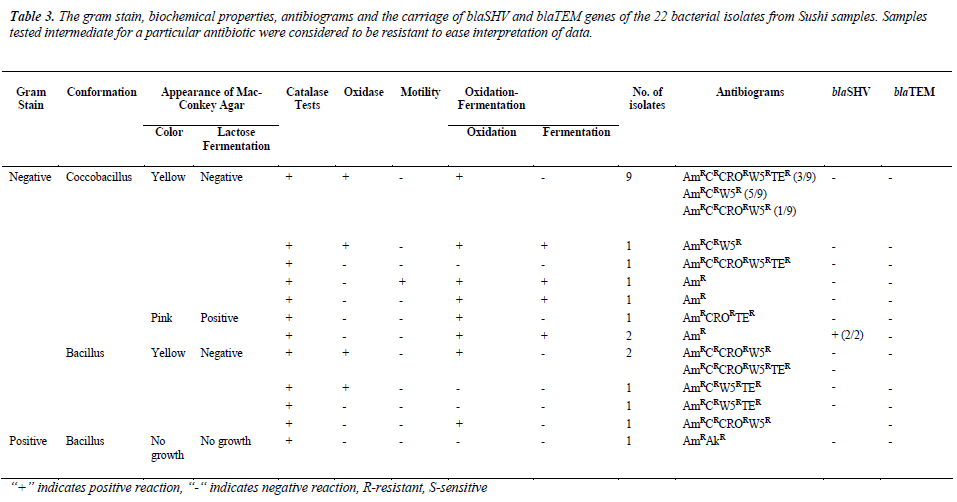
were resistant towards ampicillin and majority of the isolates were resistant against trimethoprim (72.7%) and chloramphenicol (45.5%), but susceptible towards amikacin (95.5%). Most of the isolates were resistant to at least three to five antibiotics, as tabulated in Table 2. As shown in Table 3, 13.6% (3/22) were lactosefermenters, while 81.8% (18/22) were non-lactosefermenters, whereas only 4.5% (1/22) was inhibited by the MacConkey agar. All of the bacterial isolates were catalase- positive and 63.6% (14/22) of the isolates were ox-dase-positive, while 22.7% (5/22) bacteria isolates were facultative anaerobes and 59.1% (13/22) were obligate aerobe. The remaining 18.2% (4/22) demonstrated neither oxidative nor fermentative activities. In motility test, most of the isolates were non-motile (95.5%) and all bacterial isolates were tested negative for H2S production.
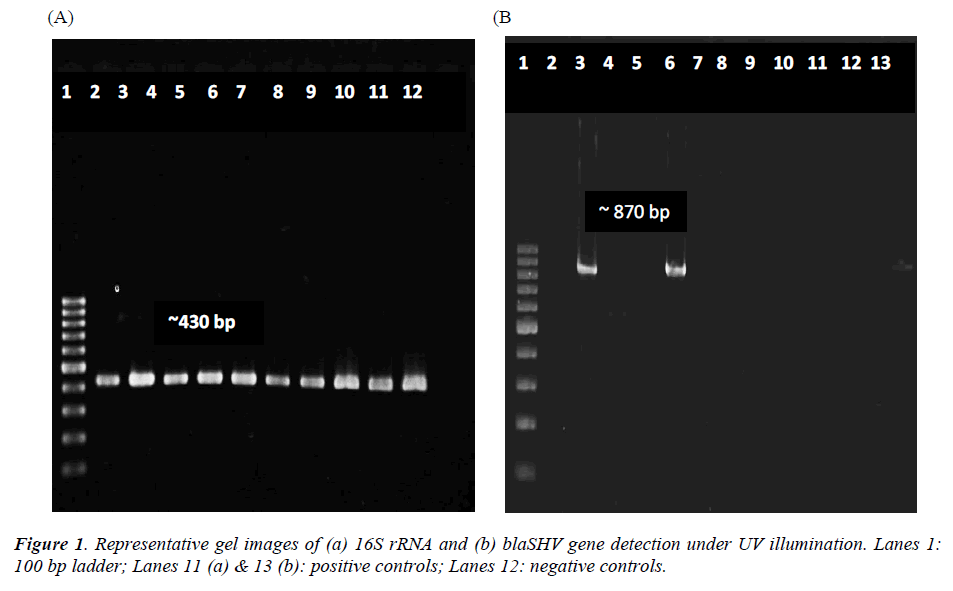
Total DNA extracted from the 22 bacterial isolates was subjected to 16S rRNA PCR amplification with gene specific primers to ensure that the DNA was intact for subsequent amplification of β-lactamases genes (Figure 1a). For blaSHV gene, two isolates (9.1%) were screened positive for presence of blaSHV (Figure 1b). DNA sequencing results proved the specificity of the PCR process to blaSHV (data not shown). For blaTEM amplification, none of the samples produced any products with the expected sizes. Since the positive control for blaTEM PCR produced a single amplicon with the expected size, this indicates the authenticity and reliability of the PCR process for blaTEM screening. Hence, none of the samples harbored any blaTEM homologous genes. Both of the bacterial isolates which carried blaSHV were subsequently identified as Raoultella ornithinolytica using bioMérieux® API 20E kit. Coincidentally, both of the isolates showed resistance to ampicillin alone (Table 3).
Discussion
In recent years, studies have demonstrated that the use of antibiotics in animals, aquaculture and most importantly, food production, is also a major factor which contributes to the emergence of antibiotic resistance in bacteria and the spread of antibiotic resistance genes [22-24]. From this study, two bacterial isolates, which were identified as R. ornithinolytica, were screened positive for presence of blaSHV. The results obtained here signified the presence of blaSHV gene in ready-to-eat sushi in Kampar which could easily transfer their antibiotic resistance genes to human. As suggested by Schjørring and Krogfelt [25], the resistant bacteria from food, such as animals, fish, plants, and vegetables that are consumed by humans can be a source of antibiotic resistant gene transfer via plasmids and transposons [23]. Indeed, a prominent example of food as a source of antibiotic-resistant bacteria in humans is the existence of the multidrug resistant extraintestinal pathogenic E. coli (ExPEC) that is responsible for many community-acquired urinary tract infections (UTIs) [26], including the outbreak of trimethoprim–sulfamethoxazole- resistant E. coli UTIs in women from the United States [27], and the community outbreak of clonally related extended-spectrum beta-lactamases (ESBLs) type CTX-M infections worldwide [28]. The sources of the strains responsible for the above outbreaks are linked to contaminated meat and other food.
The bacterial isolates which harbored blaSHV were not multidrug-resistant in their antibiogram, but they were resistant towards one particular antibiotic which is ampicillin. Nevertheless, the multi-antibiotic resistant bacteria isolates did not carry either blaSHV or blaTEM. This indicates that there could be other resistance genes other than blaTEM and blaSHV present which may be responsible for encoding the resistance, such as CTX-M. This resistance appears to have arisen through the initial transfer of the chromosomal β-lactamase gene from Kluyvera spp. to conjugative plasmids that have rapidly spread among members of the Enterobactericeae as well as other gram-negative bacteria [29]. Besides blaCTX-M, other genes related to β-lactamase have also been described. Two novel chromosomally-encoded Class-A β-lactamases, blaPLA and blaORN, from R. planticola and R. ornithinolytica strains were found to be closely related to the chromosomal β-lactamase of K. pneumoniae [30].
R. ornithinolytica is uncommon in human infections as there are only very few reported cases. Rare evidences of human infections are the reports of bacteremia caused by R. ornithinolytica in an infant [31], an 82-year old woman [32] and more recently, in cancer patients [33]. R. ornithinolytica is mostly known to cause food poisoning due to its production of histamine in scombroid fish such as mackerel, tuna and sardine [31-32]. Hence, the isolation of R. ornithinolytica from sushi in this study could imply the inadequacy of hygiene control in the restaurant or it was contaminated, either by food handlers or water sources. R. ornithinolytica inherently carries blaORN, a β-lactamase gene in its chromosome, but it is not commonly known to carry blaSHV. One other study described the presence of blaKPC, a β-lactamase gene in R. ornithinolytica which encodes for carbapenem resistance [34]. As R. ornithinolytica can easily be misidentified as Klebsiella spp., it is likely that its importance as a food contaminant, enteric pathogen and antibiotic-resistant gene reservoir may be underrated.
This study here reports the presence of blaSHV in R. ornithinolytica isolated from ready-to-eat sushi.To address antimicrobial resistance in the food supply, it is imperative that careful tracking of these foodborne associated antibiotic-resistant pathogens must be warranted in the future. It plays a critical step in understanding the link between administering antibiotics to farm animals and human illness and, ultimately to curb the development of antibiotic resistance.
Acknowledgement
The authors thank Department of Biomedical Science and UTAR for their support of this project.
References
- Amador P, Fernandes R, Prudêncio C, Brito L. Resistance to β-lactams in bacteria isolated from differenttypes of Portuguese cheese. Int J Mol Sci 2009; 10: 1538-1511.
- Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem 2009; 78: 119–146.
- Khachatourians GG (1998). Agricultural use of antibiotics and the evolution and transfer of antibioticresistant bacteria. CMAJ 1998; 159(9): 1129-1136.
- Starr MP, Reynolds DM (1951). Streptomycin resistance of coliform bacteria from turkeys fed streptomycin.Am J Public Health 1951; 41(11): 1375-1380.
- Dibner JJ, Richards JD. Antibiotic growth promoters in agriculture: history and mode of action. Poult Sci 2005;4: 634–643.
- Weese JS, Reid-Smith R, Rousseau J, Avery B. Methicillin-resistant Staphylococcus aureus (MRSA) contamination of retail pork. Can Vet J 2010; 51: 749–752.
- Van TTH, Moutafis G, Tran LT, Coloe PJ. Antibiotic resistance in foodborne bacterial contaminants in Vietnam.Appl Environ Microbiol 2007; 73(24): 7906- 7911.
- Kapperud G, Rørvik LM, Hasseltvedt V, Høiby EA, Iversen BG, Staveland K, Johnsen G, Leitao J, Herikstad H, Andersson Y, Langeland G, Gondrosen B, Lassen J. Outbreak of Shigella sonnei infection traced to imported iceberg lettuce. J Clin Microbiol 1995;33(3): 609-614.
- Hoge CW, Bodhidatta L, Tungtaem C, Echeverria P.Emergence of nalidixic acid resistance Shigella dysenteriae type 1 in Thailand: An outbreak associated with consumption of a coconut milk dessert. Int J Epidemiol 1995; 24(6): 1228-1232.
- Bassetti M, Righi E, Viscoli C. Novel β-lactam antibiotics and inhibitor combinations. Expert Opin. Investig.Drugs 2008; 17(3): 285-296.
- Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 2009;54(3): 969-976.
- Hall BG, Barlow M. Revised Ambler classification of beta-lactamases. J Antimicrob Chemother 2005; 55(6):1050-1051.
- Bali EB, Acik L, Sultan N (2010). Phenotypic and molecular characterization of SHV, TEM, and CTX-M extended- spectrum β-lactamase produced by Escherichia coli, Acinetobacter baumannii and Klebsiella isolates in a Turkish hospital. Afr J Microbiol Res 2010; 4(8): 650-654.
- Lal P, Kapil A, Das BK, Sood S. Occurrence of TEM and SHV gene in extended-spectrum β-lactamases (ESBLs) producing Klebsiella sp. isolated from a tertiary care hospital. Indian J Med Res 2007; 125: 173- 178.
- Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, Bonomo RA; International Klebsiella Study Group. (2003). Extended-spectrum betalactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob Agents Chemother 2003; 47 (11): 3554- 3560.
- Nasehi L, Shahcheraghi F, Nikbin VJ, Nematzadeh S. PER, CTX-M, TEM and SHV Beta-lactamases in clinical isolates of Klebsiella pneumoniae isolated from Tehran, Iran. Iran J Basic Med Sci 2010; 13(3): 111-118.
- Moosavian M, Deiham B. Distribution of TEM, SHV and CTX-M genes among ESBL-producing Enterobacteriaceae isolates in Iran. Afr J Microbiol Res 2012; 6(26): 5433-5439.
- Kor SB, Choo QC, Chew CH. New integron gene arrays from multiresistant clinical isolates of members of the Enterobacteriaceae and Pseudomonas aeruginosa from hospitals in Malaysia. J Med Microbiol 2013; 62: 412–420.
- Lin L, Ling BD, Li XZ. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobialresistant clinical isolates of Acinetobacter baumannii- Acinetobacter calcoaceticus complex. Int J Antimicrob Agents 2009; 33(1):27-32.
- Bert F, Branger C, Lambert-Zechovsky N (2002). Identification of PSE and OXA β-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J Antimicrob Chemother 2002;50: 11-18.
- de Oliveira Garcia D, Doi Y, Szabo D, Adams-Haduch JM, Vaz TM, Leite D, Padoveze MC, Freire MP, Silveira FP, Paterson DL. Multiclonal outbreak of Klebsiella pneumoniae producing extended-spectrum beta-lactamase CTX-M-2 and novel variant CTX-M-59 in a neonatal intensive care unit in Brazil. Antimicrob Agents Chemother 2008; 52(5):1790-1793.
- Heuer OE, Kruse H, Grave K, Collignon P, Karunasagar I, Angulo FJ. Human health consequences of use of antimicrobial agents in aquaculture. Clin Infect Dis 2009; 49(8): 1248-1253.
- Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 2011;24(4): 718-733.
- Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci USA 2013; 110(9): 3435-40.
- Schjørring S, Krogfelt KA. Assessment of bacterial antibiotic resistance transfer in the gut. Int J Microbiol 2011; 2011: 312956.
- Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, Galanakis C, Reid-Smith RJ, Tellier PP, Tellis PA, Ziebell K, Manges AR. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis 2010; 16(1): 88-95.
- Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Engl J Med 2001; 345(14): 1007-1013.
- Pitout JD, Laupland KB. Extended-spectrum β- lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 2008; 8(3): 159-166.
- Jorgensen JH, McElmeel ML, Fulcher LC, Zimmer BL. Detection of CTX-M-type Extended Spectrum β-lactamase (ESBLs) by testing with MicroScan overnight and ESBL confirmation panels. J Clin Microbiol. 2010; 48(1): 120-123.
- Walckenaer E, Poirel L, Leflon-Guibout V, Nordmann P, Nicolas-Chanoine MH. Genetic and biochemical characterization of the chromosomal class A β- lactamases of Raoultella (formerly Klebsiella) planticola and Raoultella ornithinolytica. Antimicrob Agents Chemother 2004; 48(1): 305-312.
- Mau N, Ross LA. Raoultella ornithinolytica bacteremia in an infant with visceral heterotaxy. Pediatr Infect Dis J 2010; 29(5): 477-478.
- Morais VP, Daporta MT, Bao AF, Campello MG,Andrés GQ. Enteric fever-like syndrome caused by Raoultella ornithinolytica (Klebsiella ornithinolytica). J Clin Microbiol 2009; 47(3): 868-869.
- Hadano Y, Tsukahara M, Ito K, Suzuki J, Kawamura I, Kurai H. Raoultella ornithinolytica bacteremia in cancer patients: report of three cases. Intern Med 2012; 51(22): 3193-3195.
- Castanheira M, Deshpande LM, DiPersio JR, Kang J, Weinstein MP, Jones RN. First description of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): Report from the SENTRY antimicrobial surveillance program. J Clin Microbiol 2009; 47(12): 4129-4130.