Research Article - Journal of Systems Biology & Proteome Research (2022) Volume 3, Issue 4
Ion mobility spectrometry with acquisition-based single-shot proteomics
Goutam Rout*Department of Biotechnology, Utkal University, Odisha, India
- *Corresponding Author:
- Goutam Rout
Department of Biotechnology
Utkal University, Odisha, India
E-mail: gautamrout1@gmail.com
Received: 28-Jun-2022, Manuscript No. AASBPR-22-70879; Editor assigned: 30-Jun-2022, PreQC No. AASBPR-22-70879(PQ); Reviewed: 14-Jul-2022, QC No. AASBPR-22-70879; Revised: 21-Jul-2022, Manuscript No. AASBPR-22-70879(R); Published: 28-Jul-2022, DOI: 10.35841/aasbpr-3.4.116
Citation: Rout G. Ion mobility spectrometry with acquisition-based single-shot proteomics. J Syst Bio Proteome Res. 2022;3(4):116
Abstract
Ion mobility spectrometry, a method that divides gas phase ions by their size and shape and may be hybridised with traditional LC and MS, has recently gained popularity thanks to improvements in efficiency and simplicity of use. Here, we examine the most recent advancements in Trapped Ion Mobility Spectrometry (TIMS) in conjunction with TOF mass analysis. Particularly in terms of sequencing speed and sensitivity, the Parallel Accumulation-Serial Fragmentation (PASEF) operation mode offers exceptional benefits. Its distinguishing characteristic is that it coordinates the downstream selection of precursors for fragmentation in a TIMS quadrupole TOF configuration with the release of ions from the TIMS device. When doing data-dependent or focused analysis, the number of peptide fragment ion spectra obtained decreases as ions are compressed into small ion mobility peaks data-dependent or targeted analyses can be increased by an order of magnitude without compromising sensitivity. The PASEF concept increases the effectiveness of data-independent collection by taking advantage of the correlation between ion mobility and mass. This makes the method suitable for ultrasensitive measurements down to single cells as well as quick proteome profiling, a feature that is becoming more and more significant in clinical proteomics. The development of neural networks that can predict collisional cross section values solely based on peptide sequences is made possible by the speed and accuracy of TIMS and PASEF. PASEF also allows for precise measurements of collisional cross section values at the scale of more than a million data points. For isobaric sequences or positional isomers of post-translational modifications, the values of the peptide collisional cross section can vary. This additional data may be used to guide data collecting in real time or to boost the accuracy of peptide identifications after post-processing. Due to these advancements, TIMS quadrupole TOF PASEF is now a strong and flexible platform for proteomics and other fields.
Keywords
Single-shot proteomics, DIA-MS, FAIMS, Feces, Depression.
Introduction
Genes produce molecules called proteins, which are multifunctional molecules. Because proteins are generated from messenger RNA, protein expression prediction using transcriptome analysis is commonly used, even though it is ideal to evaluate the protein accumulation level by measuring the protein itself (mRNA). This is frequently linked to the fact that transcriptome analysis can identify more compounds than proteome analysis [1]. The significance of detecting the protein numbers itself in the multiomics era is undoubtedly increased by reports showing the correlation between mRNA and protein expression levels is only moderately high. The depth and ease of proteome analysis therefore need to be further improved. Quadrupole (Q)-time of flight-MS or Q-Orbitrap MS in the DIA-MS is generally used [2]. Most of the proteins reported in studies of over proteins discovered by DIA-MS are Q-Orbitrap MS. The molecules that pass through Q in the case of Q-Orbitrap MS are broken up, and the ions are gathered by the ion routing multipole and identified by Orbitrap MS. The Q-Orbitrap MS makes it possible to measure trace compounds with a high analytical depth thanks to this accumulation by the ion routing multipole [3]. The front end of Q-Orbitrap MS can now be equipped with high-field asymmetric waveform ion mobility spectrometry, which lowers the amount of chemical background noise and ion complexity during ion mobility separation. It is anticipated that FAIMS will increase protein identifications.
In order to increase the analytical depth of the straightforward single-shot proteome analysis, the parameters of FAIMS were tuned for DIA-MS in this study. Furthermore, using a mouse model of Recurrent Social Defeat Stress (R-SDS), our approach was used to analyse the faecal proteome and identify changes in faecal host proteins as a result of sadness [4]. Two behavioural tests-the elevated plus-maze test and the social interaction test-were performed on the defeated and control mice. All mice were habituated to the behavioural chamber before receiving R-SDS before the social interaction test. After R-SDS, the defeated and control mice were put in the chamber with a novel ICR mouse that had been placed at one end of the chamber and was encased in a metal meshwork.
The chamber was open for the experimental mice to freely wander for 150 seconds. An avoidance zone was established on the other side of the metal meshwork, and the amount of time spent there was used as a gauge of depressive-like behaviours. For the elevated plus-maze test, two closed arms and two open arms of the behavioural maze equipment, which included two open arms and two closed arms, were put on the ends of the defeated and control mice [5]. After five minutes of unrestricted maze exploration, the mice were placed in open arms to measure their anxiety levels. Reduced time in the open arms is regarded as increased worry.
The requirement for proteome analysis with high proteome coverage in single-shot measurements has arisen as a result of the development of Mass Spectrometry (MS) and analytical methodologies. Ion mobility spectrometry and Data- Independent Acquisition (DIA)-MS have received attention as methods for in-depth proteome investigation. With the help of DIA-optimized MS's high-field asymmetric waveform ion mobility spectrometry settings, we sought to increase the proteome coverage through single-shot observations [6]. Within 120 minutes of the MS measurement time, our existing proteome analysis method detected more than protein groups from cell digests. Furthermore, using our method to analyse host proteins found in mouse faeces, we found as many as 892 host protein groups in a mouse model of Repeated Social Defeat Stress (R-SDS), a tool used to study depression. It's interesting to note that 285 proteins that R-SDS increased were connected to mental illnesses [7]. We may be able to grasp the pathologies of mental illness non-invasively with the use of the faecal host protein profiling by deep proteome analysis. Therefore, because our method enables the investigation of large proteome coverage in a single-shot measurement, it will be useful for a detailed comparison of protein expression levels for biological and medical research.
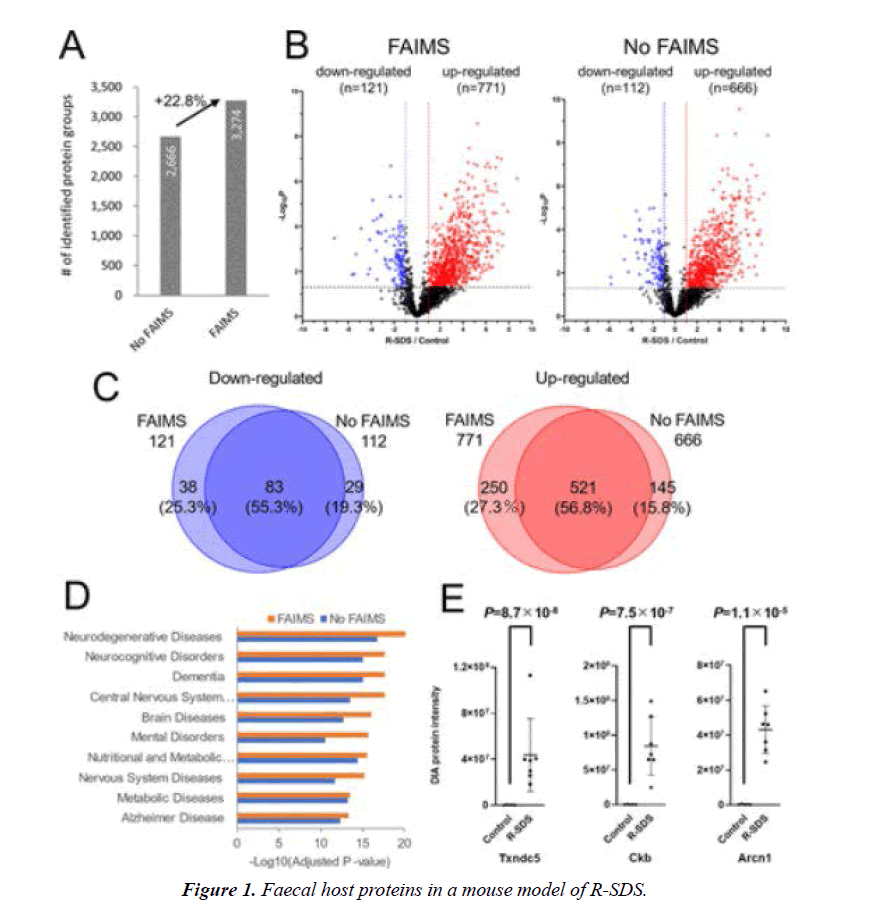
The ultra-deep proteome analysis revealed changes in faecal host proteins in a mouse model of R-SDS (Figure 1). Using DIA-MS with and without FAIMS, the number of protein groups detected is shown in (A). (B) Volcano plot of the host protein concentrations from the faeces of control and R-SDS model mice. In the faeces of the R-SDS model mice, the red dots represented proteins that were elevated, and the blue dots represented proteins that were downregulated [8]. (C) Comparison of the proteins in R-SDS faeces that were altered and those that were not, as determined by DIA-MS with FAIMS and DIA-MS without FAIMS. (D) Analysis of host proteins that were overexpressed in the faeces of R-SDS model mice using disease ontology enrichment [9]. (E) Protein intensities of three representative proteins upregulated in the feces of the R-SDS model mice.
Liquid Chromatography
Fundamental physics dictates that stationary ions submerged in a gas flow follow the same rules as those that apply to ions drifting through a stationary gas (as in a drift tube IMS).
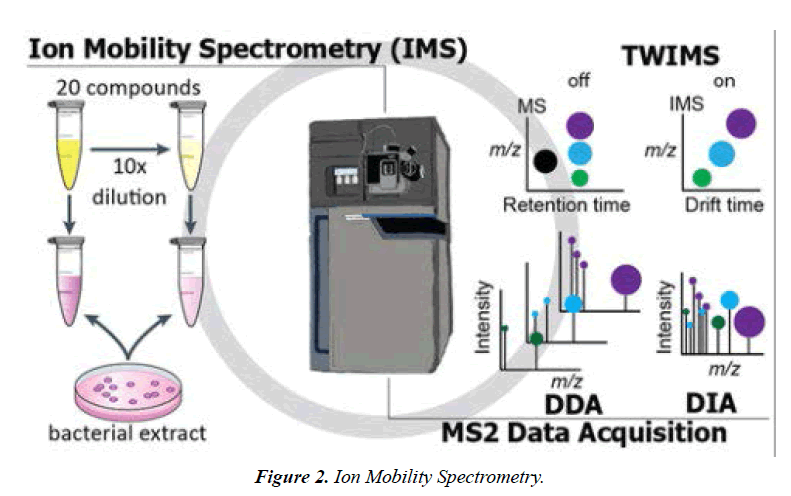
To put this idea into practise, the TIMS traps ions in an electrodynamic tunnel that directs a gas flow from the entry to the exit at a pressure of roughly 2 to 3 mbar. However, the drag is dependent on the ions' cross section; a higher cross section results in a stronger drag. The drag force on the ions caused by the gas flow is essentially constant along the length of the device Figure 2.
An analytical Direct Current (DC) field that strengthens down the tunnel opposes this drag force. Ions entering the TIMS tunnel come to rest at an axial location where the force from the analytical field balances the drag force on the ions (directed toward the tunnel exit) (directed toward the tunnel entrance). Low-mobility ions are trapped higher up the electric field gradient and farther into the TIMS tunnel because they need stronger fields to offset the drag from the gas. Highmobility ions, on the other hand, settle closer to the opening [10]. Further ions are stopped from entering the tunnel once the desired accumulation time has been reached, and mobility analysis starts. As the analytical DC field intensity is gradually decreased, ions are eluted based on their mobility during the analysis [11]. The tunnel entrance potential is ramped at a consistent and user-defined rate while the exit potential is kept fixed. However, since the entry potential may be arbitrarily changed, arbitrary scanning routines can theoretically be programmed. The majority of the analytical separation is accomplished at the electric field plateau as ions are released from the TIMS device, according to simulations of ion dynamics and numerical modelling.
A single TIMS device cannot use more than half of the ions produced by a continuous ion source, assuming equal accumulation and ramp times [12]. Extended the ion tunnel and divided it into a trap, a transfer area, and a TIMS analyzer in order to get over this restriction. The roles of gathering ions and analysing ions are spatially separated in this "dual TIMS" design. As a result, ions can be gathered and examined simultaneously in time. Ions are gathered and detained in the trap section before being swiftly transmitted to the TIMS analyzer, where their mobility is then examined. The subsequent batch of ions, such as are simultaneously added to the trapping portion [13].
Achieving the requisite ion mobility resolution and filling the TIMS device to capacity can both be accomplished in practise with an acquisition time of roughly 100ms. Tandem TIMS is a more recent development. In this setup, anion gate is used to pass particular ion mobility ranges to the second TIMS analyser [14]. This makes it possible to conduct collisional activation tests on species that have been chosen based on mobility at the interface, for instance, to study protein unfolding or to do more research on fragment ions in the downstream TIMS. Thus, tandem TIMS is particularly interesting for top-down proteomics, especially if native-like protein structures are conserved, but it can also be used to analyse peptides and small molecules in a similar way [15].
All precursor ions are simultaneously gathered in the TIMS device during MS/MS studies using a TIMS-QTOF mass spectrometer, and they are then released during the course of TIMS scans or "frames" that typically last 100ms in narrow ion mobility peaks of around 2ms (full width at half maximum). When compared to continuous capture without TIMS, this provides the immediate benefit of increasing the signal-tonoise ratio because the signal is amplified by more than one order of magnitude. (Take note that the detection system has new issues as a result; the concentrated ion packets may get saturated). Additionally, coeluting peptides of comparable mass that are interfering background ions are separated from by ion mobility, increasing the relative fraction of fragment ions coming from the targeted precursor in the MS/MS spectrum [16]. In agreement with this, Ogata and Ishihama revealed that, although TIMS is now limited to 1 Da-spaced reporter ions due to the instrument's mass resolution, it can at least partially solve the ratio-compression issue of tandem mass tag-based measurement.
Conclusion
By optimising the FAIMS parameters in DIA-MS, the proteome coverage in single-shot proteome analysis was increased. Furthermore, utilising DIA-MS with FAIMS for deep proteome analysis, we were able to find over 10,000 protein groups, which is more than the number of genes found by RNA-seq. We were able to identify 892 variable proteins via R-SDS, a mouse model to research depression, using our 10K proteome technique to host protein analysis in mouse faeces. Therefore, our 10K proteome approach's characterization of faecal host proteins may enable us to gain a non-invasive understanding of the pathology of a number of disorders linked to gastrointestinal changes.
References
- Buccitelli C, Selbach M, mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet. 2020;21:630-44.
- Nagaraj N, Wisniewski JR, Geiger T, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Molecular systems biology. 2011;7(1):548.
- Horie K, Kamakura T, Ikegami T, et al. Hydrophilic interaction chromatography using a meter-scale monolithic silica capillary column for proteomics LC-MS. Analytical chemistry. 2014;86(8):3817-24.
- Kawashima Y, Watanabe E, Umeyama T, et al. Optimization of data-independent acquisition mass spectrometry for deep and highly sensitive proteomic analysis. Int J Mol Sci. 2019;20(23):5932.
- Sielaff M, Kuharev J, Bohn T, et al. Evaluation of FASP, SP3, and iST protocols for proteomic sample preparation in the low microgram range. J Proteome Res. 2017;16(11):4060-72.
- Searle BC, Swearingen KE, Barnes CA, et al. Generating high quality libraries for DIA MS with empirically corrected peptide predictions. Nat Commun. 2020;11(1):1-10.
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44-57.
- Demichev V, Messner CB, Vernardis SI, et al. DIA-NN: Neural networks and interference correction enable deep coverage in high-throughput proteomics. bioRxiv. 2018:282699.
- Naylor CN, Ridgeway ME, Park MA et al. Evaluation of trapped ion mobility spectrometry source conditions using benzylammonium thermometer ions. JASMS. 2020;31(7):1593-602.
- Morsa D, Hanozin E, Eppe G, et al. Effective temperature and structural rearrangement in trapped ion mobility spectrometry. Anal Chem. 2020;92(6):4573-82.
- Adachi J, Hashiguchi K, Nagano M, et al. Improved proteome and phosphoproteome analysis on a cation exchanger by a combined acid and salt gradient. Anal chem. 2016;88(16):7899-903.
- Li KW, Gonzalez-Lozano MA, Koopmans F, et al. Recent developments in data independent acquisition (DIA) mass spectrometry: Application of quantitative analysis of the brain proteome. Front Mol Neurosci. 2020;13:564446.
- Meier F, Geyer PE, Virreira Winter S, et al. BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat Methods. 2018;15(6):440-48.
- Nie X, Kitaoka S, Tanaka K, et al. The innate immune receptors TLR2/4 mediate repeated social defeat stress-induced social avoidance through prefrontal microglial activation. Neuron. 2018;99(3):464-79.
- Dinan TG, Cryan JF. Regulation of the stress response by the gut microbiota: Implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37(9):1369-78.
- Knudsen JK, Bundgaard‐Nielsen C, Hjerrild S, et al. Gut microbiota variations in patients diagnosed with major depressive disorder-A systematic review. Brain Behav. 2021;11(7):e02177.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref