Research Article - Biomedical Research (2017) Volume 28, Issue 6
Investigation of the relationship between class-1 integrons and per-1 enzyme in ceftazidime resistant Pseudomonas aeruginosa
Ayşegül Opuş1, Tuba Yıldırım2*, Asuman Birinci3 and Belma Durupınar31Microbiology and Clinical Microbiology Laboratory, Konya Research and Education Hospital, Konya, Turkey
2Department of Biology, Faculty of Arts and Sciences, Amasya University, Amasya, Turkey
3Department of Medical Microbiology, Faculty of Medicine, Ondokuz Mayıs University, Samsun, Turkey
- *Corresponding Author:
- Tuba Yıldırım
Department of Biology, Faculty of Arts and Sciences
Amasya University, Turkey
Accepted date: November 09, 2016
Abstract
Pseudomonas aeruginosa strains, especially multidrug resistant, have great of importance among nosocomial infection isolates. Production of beta lactamase is an important mechanism in gram-negative bacteria for resistance to beta-lactam antibiotics. PER-1 enzyme is derived from an extended spectrum beta-lactamase that is non-TEM, non-SHV-derived in class A and especially causes ceftazidime resistance. In this study, our aim was to investigate the relationship between CLASS-1 integrons and PER-1 enzyme in ceftazidime-resistance Pseudomonas aeruginosa. The PER-1 type beta lactamase enzyme that causes ceftazidime-resistance, determines the frequency, and detects the relationship between the enzyme and CLASS-1 integrons by PCR in 100 (one hundred) ceftazidime-resistant Pseudomonas aeruginosa (P. aeruginosa) isolated at Ondokuz May?s University Hospital (Turkey) between 2007 and 2008. In this study, blaPER-1 was detected in 40% (40/100) of the isolates. Four principal clones, which were detected in P. aeruginosa strains were responsible for high prevalence using Random Amplified Polymorphic DNA-PCR (RAPD-PCR) methods. CLASS-1 integron was detected in 62.5% (25/40) of the PER-1 enzyme bearing strains and association between blaPER-1 and CLASS-1 integrons were shown in 2 (two) Pseudomonas aeruginosa strain. Also resistance rates of PRL antibiotic in blaPER-1 negative group was found to be significantly higher against blaPER-1 positive group, resistance rates of other antibiotics were no different between these two groups. We concluded that PER-1 enzyme is common in our hospital and their clonal diversity indicates horizontal dissemination, the association between bla PER-1 and CLASS-1 integrons can accelerate dissemination of this gene.
Keywords
Beta-lactamase, PER-1, Pseudomonas aeruginosa, Class-1 integron.
Introduction
Pseudomonas aeruginosa is one of the most important nosocomial pathogens that causes nosocomial infections and frequently leads to mortality and morbidity [1,2]. P. aeruginosa infections are often difficult or impossible to eradicate. To some extent, this is due to high-level resistance to many antimicrobial and disinfectants as a result of both intrinsic and acquired mechanisms [3,4]. Several mechanisms can contribute to acquire beta-lactam resistance in P. aeruginosa, including beta-lactamase production, the upregulation of efflux systems, and decreased outer membrane permeability [5,6]. Betalactamases are the most common cause of bacterial resistance to beta-lactam antimicrobial agents [2]. Among the acquired enzymes, PER-1 is class A extended-spectrum beta-lactamase (ESBL) of notable clinical importance due to its high level of activity especially causes ceftazidime and to the broad diffusion achieved by it in some epidemiological settings [5,7-9]. Various classes of ESBLs (A, B and D) have been found recently in P. aeruginosa. Five types of class A ESBLs (PER, VEB, GES, IBC, TEM and SHV) were recently reported in P. aeruginosa; however, these were found in limited regions [10]. The PSE, OXA and PER types are the most common ESBLs found in P. aeruginosa isolates [10-13]. The PER-1 and VEB-1 types belong to class A enzymes and relate to a high level of resistance to cephems, monobactams and ceftazidime [12-14].
The resistance genes such as blaPER-1 can be transferred among bacteria by mobile genetic elements, such as integrons and even ESBL located on integron and facilitate these elements [15-17]. Integrons structure contains essential elements for insertion and mobilization of gene cassettes generally possess antimicrobial resistance genes [18,19]. In particular, much research effort has been devoted to integrons belonging to classes 1, 2 and 3, the classes most commonly associated with the spread of antibiotic resistance in pathogens [20]. Class 1 integron is composed of a 5'- conserved segment (5'CS) including the integrase gene intI and attI genes and Pant promoter and 3'-conserved segment (3'CS) [21,22]. Class-1 integrons, which are most commonly found in antimicrobialresistance clinical isolates, possess two conserved segments located on either side of the integrated genes [16-23] and have a threatening potential for the development of antimicrobial resistance and the emergence of MDR profiles in clinical bacteria, gram negative bacteria as well as P. aeruginosa [18]. To date the first report to analyse blaPER-1 has been shown to be related with these genetic determinants [6-15]. The objective of this study was to determine the PER-1 type betalactamase enzyme that causes ceftazidime-resistance P. aeruginosa using molecular methods. Then, we would be able to determine the frequency and detect the relationship between the enzyme and class-1 integrons.
Materials and Methods
Clinical isolates
One hundred randomly selected strains among one hundred seventy-nine (N=179) strains of P. aeruginosa were analysed in this study. These strains were nonreplicate clinical isolates from hospitalized patients in Ondokuz Mayıs University Hospital (Turkey) during the period 2007 and 2008. All isolates were resistant to ceftazidime. P. aeruginosa ATTC 27853 was used as the reference strain for Kirby-Bauer disk diffusion method.
Susceptibility testing and screening for ESBLproducing strains
In vitro susceptibility was determined by a Kirby-Bauer disk diffusion method on Mueller-Hinton (MH) Agar (Merck, Darmstadt, Germany) plates as recommended by the “Clinical and Laboratory Standarts Institute” (CLSI) [24]. The results of susceptibility testing were interpreted according to the criteria of the CLSI. Antimicrobial agents were purchased from Oxoid (Chemical Co, Hampshire, England). The double-disk synergy test (DDST) was performed with cefotaxime, ceftazidime, aztreonam, and amoxicillin/clavulanic acid disks on Mueller- Hilton agar plates, and the results were interpreted as described previously [25].
Determination of ceftazidime MIC values
Ceftazidime MIC values of the blaPER-1 positive strains were determined by microdilution method according to CLSI criteria.
Detection of blaPER-1
Total DNA was extracted from the P. aeruginosa isolates as described previously by phenol chloroform method [26]. Primers (Tib Molbiol, Berlin, Germany) were PERA, 5’-ATG AAT GTC ATT ATA AAAGC-3’, and PERD, 5’- AAT TTG GGC TTA GGG CAGAA-3’, yielding a 926-bp product. blaPER-1 was sought among the isolates by PCR assay according to [1]. Amplification reactions were performed in 50 μL including 2 μL of total DNA (50 ng) as template, 2.5 mM of MgCl2, 2 mM dNTP, 5U of Hot Start Taq DNA polymerase (Fermentas, Vilnius, Lithuania), and 10 pmol of primers. PCR conditions were 5 min at 94°C, followed by 35 cycles of 45 sec at 95°C, 45 sec at 56°C and 1 min at 72°C, followed by a final extension for 7 min at 72°C. The MultiBlock PCR System (Thermo, CA, USA) was used for gene amplification. Electrophoresis was carried out in conventional 1X Tris- Borate-EDTA (TBE) buffer (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA, pH: 8.4) with 1% agarose. 1 kb DNA ladder mix (Fermentas, Vilnius, Lithuania) was used to provide molecular size markers. PCR products were loaded onto 2% agarose gel containing 1X TBE and stained with 5 μg/ml ethidium bromide (Sigma, CA, USA). A 100 bp DNA Ladder Plus (Fermentas, Vilnius, Lithuania) was used to provide molecular size markers, and photographed using a Gel/ ChemiDoc XRS system (Bio-Rad Laboratories, CA, USA).
Molecular fingerprinting
The clonal relatedness of the PER-1 positive isolates was investigated by RAPD-PCR analysis with the ERIC-2 primer (Tib Molbiol, Berlin, Germany) 5’-AAG TAA GTG ACT GGG GTG AGC G-3’ [3] and an amplification protocol: 1 min at 95°C, followed by 30 cycles of 1 min at 94°C, 1 min at 40°C, and 1 min at 72°C, followed by a final extension for 5 min at 72°C. PCR products were loaded onto 2% agarose gel containing 1X TBE and stained with 5 μg/ml ethidium bromide (Sigma, CA, USA) observed using a Gel/ChemiDoc XRS system. 1 kb DNA ladder mix (Fermentas, Vilnius, Lithuania) was used to provide molecular size markers. To evaluate similarity between the strains, Jaccard coefficients [SJ=nAB × 1/ (nAB + a + b)] were calculated according to the banding patterns (for symbol explanation and further details see Soll 2000). When the calculated SJ was <0.8, two isolates were accepted as “clonally distinct”. Isolates were classified as “clonally related” when the calculated value was 0.8 SJ<1.0; when it was 1.0, the isolates were considered as “identical”.
Integron PCR
PCR was performed in a final volume of 50 μL containing 5 μL of total DNA (50 ng) as template, 3 mM of MgCl2, 200 μM dNTP, 5U of Taq DNA polymerase (Fermentas, Vilnius, Lithuania), and 100 pmol of 3’ -CS 5’- AAG CAG ACT TGA CCT GA-3’ primer and 5’-CS 5’- GGC ATC CAA GCA GCA AG-3’ primer (Tıb Molbiol, Berlin, Germany). Amplification reactions consisted of denaturation at 95°C for 10 min followed by 30 cycles of denaturation at 94°C/1 min, annealing at 55°C/1 min and elongation at 72°C/5 min, followed by a final extension for 4 min at 72°C. Amplification products were analysed using 1% agarose gel electrophoresis (1X TBE buffer at 900 mA), stained with ethidium bromide solution and visualized using a Gel/ChemiDoc XRS system. 1 kb DNA ladder mix was used to provide molecular size markers.
Investigation of blaPER-1 location in class-1 integrons
Integron location of blaPER-1 was investigated by PCR. A combination of primer 5’-CS or 3’-CS and a primer (PERA, PERD) specific for the blaPER-1 gene were also used to identify the location of blaPER-1 in the integrin [19]. The extension step was increased to 6 min, and the amount of Taq DNA polymerase was increased to 6 units per reaction mixture to amplify large DNA fragments [15,27,28].
Statistical analyses
Representatives of each RAPD-PCR pattern were used integron-PCR experiments. Dendrograms were formed using Jaccard coefficients and dendogram’s Biomerics version 5.10 with UPGMA (Unweighted pair group method with mathematical averaging) agglomerative hierarchical cluster method for PER-1 producing P. aeruginosa strains. Phylogenetic analysis program PAUP (ver. 4.0b10) was also used for implementing the UPGMA method. Pearson chisquare test was used for comparison of PER-1 enzyme including (blaPER-1) and non-including groups about in terms of observed resistance rates of aztreonam, cefepime, cefotaxime, piperacillin, piperacillin-tazobactam, imipenem and meropenem. p<0.05 was taken for statistical level of significance and PASW (SPSS, ver. 18) was used for the calculation of descriptive statistics and making chi-square tests.
Results
A total of 179 nonrepetitive ceftazidime-resistant P. aeruginosa isolates were collected from hospitalized patients in Ondokuz Mayıs University Hospital (Turkey) between 2007 and 2008. Ceftazidime resistance accounted for 27% (179/641) of all the P. aeruginosa isolates. According to the disk diffusion susceptibility test results, the resistance rates of P. aeruginosa isolates to aztreonam, cefepime, cefotaxime, piperacillin, piperacillin-tazobactam, imipenem and meropenem was found 100% (100/100), 88% (88/100), 100% (100/100), 67% (67/100), 43% (43/100), 75% (75/100) and 70% (70/100) respectively. Multiple antimicrobial resistance phenotypes according to blaPER-1 occurrences are shown in (Table 1). In this study, the percentage of PER-1 of ceftazidime resistant P. aeruginosa strains was found 40% (40/100) similar to other studies. Difference between blaPER-1 positive and negative strains in terms of resistance ratio of seven antibiotics were compared and only resistance rate of PRL antibiotic significantly higher than any other blaPER-1 negative group (p=0.014). This result was not statistically significant.
| blaPER (+) resistance rate% (n=40) | blaPER (-) resistance rate% (n=60) | χ2 with 1 df | P | |||
|---|---|---|---|---|---|---|
| Antimicrobial agent | Number | % | Number | % | ||
| TZP | 17 | 42.5 | 27 | 45 | 0.061 | 0.805 |
| FEP | 38 | 95 | 54 | 90 | 0.815 | 0.367 |
| PRL | 16 | 40 | 39 | 65 | 6.06 | 0.014 |
| CTX | 40 | 100 | 60 | 100 | 0 | 1 |
| MEM | 24 | 60 | 35 | 58.5 | 0.028 | 0.868 |
| ATM | 40 | 100 | 60 | 100 | 0 | 1 |
| IPM | 28 | 70 | 41 | 68.3 | 0.031 | 0.86 |
| TZP: piperacillin-tazobactam; FEP: cefepime; PRL: piperacillin; CTX: cefotaxime; MEM: meropenem; ATM: aztreonam; IPM: imipenem | ||||||
Table 1. Antibiotic resistance phenotypes of blaPER-1, positive and negative isolates.
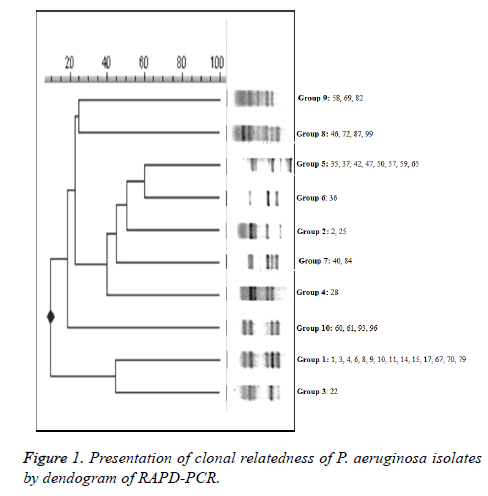
Twenty out of the 40 PER-1 producing P. aeruginosa isolates were found positive for ESBL by double disk diffusion method. DDST standardize were not done for Pseudomonas and ESBL conjectural only the interpretation can be done. The tested isolates were divided into 10 groups according to their banding patterns by RAPD-PCR (Figure 1). Group 1 (n=14), Group 5 (n=8), Group 8 (n=4), Group 10 (n=4) major clones, as well as several strains containing the sub-groups were determined by RAPD-PCR. We showed that four principal clones that were detected in P. aeruginosa strains were responsible for high prevalence by RAPD-PCR results. In our data, it has been detected that 22 of P. aeruginosa strains (55%) attended two different patterns and these main patterns included strains especially that isolated from urine and endotracheal aspirates from patients admitted in the urology department and adult intensive care unit. Group 1 (n=14) were isolated from urine (n=9), catheter (n=1), wound (n=1), serebro spinal fluid (n=1), tracheal aspirate (n=2), taken from urology and intensive care wards. Group 5 (n=8) were isolated from urine (n=3), tracheal aspirate (n=3), wound (n=1), sputum (n=1) taken from urology and intensive care wards. Group 2, 3, 4, 6, 7 and 9 consisted of 10 isolates were from various wards and specimens. Thirty nine (45.3%) isolates were detected as multi-drug resistant. PCR results showed that 37 (43%) of P. aeruginosa isolates harbored the class 1 integron and of these 27 (69.2%) isolates were multidrug-resistant.
CLASS-1 integron was detected in 62.5% (25/40) of the PER-1 enzyme bearing strains. P. aeruginosa isolates showed 3-4 banding patterns between 500-4000 bp in length. This result indicated that there were more than one class-1 integron according to number and size. Therefore, we evaluated them in three groups A, B and C in terms of band pattern. PER-1 producing P. aeruginosa strain’s ceftazidim MIC values were ≥ 64. Ceftazidim MIC values were different in integron (+) and (-) isolates, MIC values of integron positive isolates were higher (Table 2). The relationship between blaPER-1 and CLASS-1 integron was also examined with PCR. We obtained PCR product more than 1 kb in three P. aeruginosa isolates. These three isolates were used as a template for PCR by using PERA and PERD primers to understand whether they consist of PER-1 gene or not. We obtained PER-1 specific PCR product (900 bp) in only two isolates. We concluded that PER-1 enzyme is common in our hospital and clonal diversity indicates horizontal dissemination, the association between blaPER-1 and CLASS-1 integrons can accelerate dissemination of this gene. In the past years there were many studies that showed PER-1 enzyme was carried by transposons and plasmids [28,29] but at the present day there was only one study that demonstrated PER-1 enzyme carried by CLASS-1 integron [15].
| Number of isolates | RAPD profile | Service | Specimen | Integron pattern | Ceftazidim MIC values |
|---|---|---|---|---|---|
| 1 | R1 | Urology | Urine | A | 1024-R |
| 3 | R1 | Urology | Urine | A | 512-R |
| 4 | R1 | Urology | Wound | A | 1024-R |
| 6 | R1 | Urology | Urine | A | 128-R |
| 8 | R1 | Intensive care | CSF | A | 128-R |
| 9 | R1 | Urology | Urine | A | 256-R |
| 10 | R1 | Intensive care | Catheter | A | 512-R |
| 11 | R1 | Urology | Urine | A | 128-R |
| 14 | R1 | Intensive care | Tracheal aspirate | A | 512-R |
| 15 | R1 | Urology | Urine | A | 512-R |
| 17 | R1 | Intensive care | Urine | A | 256-R |
| 67 | R1 | Urology | Urine | A | 64-R |
| 70 | R1 | Urology | Urine | A | 512-R |
| 79 | R1 | Urology | Urine | A | 128-R |
| 25 | R2 | Intensive care | Pleural fluid | Negative | 64-R |
| 2 | R2 | Internal medicine | Urine | Negative | 64-R |
| 22 | R3 | Intensive care | Tracheal aspirate | Negative | 128-R |
| 28 | R4 | Urology | Urine | Negative | 64-R |
| 35 | R5 | Urology | Urine | B | 64-R |
| 42 | R5 | Intensive care | Tracheal aspirate | B | 128-R |
| 37 | R5 | Cardiovascular surgery | Wound | B | 512-R |
| 47 | R5 | Urology | Urine | B | 128-R |
| 50 | R5 | Neurology | Sputum | B | 256-R |
| 57 | R5 | Urology | Urine | B | 512-R |
| 59 | R5 | Intensive care | Tracheal aspirate | B | 256-R |
| 65 | R5 | Intensive care | Tracheal aspirate | B | 256-R |
| 36 | R6 | Cardiovascular surgery | Wound | Negative | 64-R |
| 40 | R7 | Pediatrics | Wound | Negative | 128-R |
| 84 | R7 | Neurosurgery | Urine | Negative | 128-R |
| 46 | R8 | Internal medicine | Urine | C | 256-R |
| 72 | R8 | Urology | Urine | C | 128-R |
| 87 | R8 | Pediatrics | Tracheal aspirate | C | 256-R |
| 99 | R8 | Urology | Urine | C | 256-R |
| 58 | R9 | Pediatrics | Tracheal aspirate | Negative | 64-R |
| 69 | R9 | Internal medicine | Sputum | Negative | 64-R |
| 82 | R9 | Pediatrics | Urine | Negative | 128-R |
| 60 | R10 | Neurosurgery | Urine | Negative | 128-R |
| 61 | R10 | Neurosurgery | Urine | Negative | 64-R |
| 93 | R10 | Neurosurgery | Tracheal aspirate | Negative | 64-R |
| 96 | R10 | Neurosurgery | Tracheal aspirate | Negative | 256-R |
Table 2. RAPD-PCR and integron patterns of blaPER-1 (+) P. aeruginosa isolates.
It has been shown that the P. aeruginosa strains that have different RAPD-PCR patterns have different integrons. We detected that in the two strains that were group representative blaPER-1 have relationship with CLASS-1 integrons. It would be necessary to support this relationship with DNA sequence analyses.
Discussion
The aim of this study was to determine the PER-1 type betalactamase enzyme that causes ceftazidime-resistance P. aeruginosa using molecular methods. Then, we would be able to determine the frequency and detect the relationship between the enzyme and class-1 integrons. Ceftazidime-resistance rates of 18-36% have been reported previously in P. aeruginosa isolates from hospitalized patients in Turkey [30-32]. Resistance to piperacillin, piperacillin-tazobactam, and other major antibiotics was found to be extremely high, which is similar to previous studies in Turkey [30-32]. Kolaylı et al. [32] reported the PER-1 presence in 55.4% of ceftazidimeresistant P. aeruginosa strains, isolated in ICUs of seven university hospitals. Moreover, 86% ceftazidime-resistant ICU (Turkey) isolated strains of P. aeruginosa were found to be PER-1 positive [15-32] and found that the percentage of PER-1 of ceftazidime resistant P. aeruginosa strains was 46.2%. In addition, [33] also reported that among the 67 ESBL-positive strains, 50 (74.62%), 33 (49.25%) and 21 (31.34%) were positive for OXA-10, PER-1 and VEB-1 genes, respectively, indicating high-level resistance to penicillins, CAZ and CTX.
We also perfomed double disk synergy test but this test fail to detect strains that produce PER-1 enzymes [30,34]. Therefore, we also carried out PCR to confirm the detection of P. aeruginosa producing PER-1 enzymes. P. aeruginosa strains that have different RAPD-PCR patterns have blaPER-1 and it has been horizontally disseminated. Banding patterns were evaluated in three groups as A, B, C. PER-1 producing P. aeruginosa strains ceftazidim MIC values were ≥ 64. Ceftazidim MIC values were different in integron (+) and (-) isolates. MIC values of integron positive isolates were higher. These results were similar to other studies which investigated presence and frequency of integrons [23,35,36]. In the past years there were many studies that showed PER-1 enzyme was carried by transposons and plasmids [28,29] and one of them demonstrated PER-1 enzyme carried by CLASS-1 integron [15]. A significant correlation was obtained between the presence of integrons and resistance against imipenem, ceftazidime, piperacillin and ciprofloxacin [18]. CLASS-1 integrons were detected in 27.3% of the multi-drug resistant P. aeruginosa isolates. Their results suggested the relationship between CLASS-1 integron and antibiotics resistance [37].
In another study by Martinez et al. [20], they studied with environmental and clinical isolates of P. aeruginosa to identify presence and spread of class 1 integrons and even characterise their variable regions. For this purpose, they examined a total of 76 isolates and PCR was used for integron detection. PCR detected class 1 integrons in 26 of the 56 clinical isolates. Similar to our study, they also searched 3’CS and 5’CS sequences. Iraj et al. investigated frequency of class 1 integrons between multidrug resistance and non MDR strains from burn wound samples by using PCR [18]. They concluded that 43% had class 1 integrons. The presence of integrons can be effective in antibiotic resistance of Pseudomonas aeruginosa cases. To support this opinion, Kouchaksaraei et al. showed that the resistance of integron positive strains to gentamicin, ticarcillin, ciprofloxacin, imipenem, amikacin, ceftriaxone, ofloxacin, ceftazidime and cefepime antibiotics was 81.9%, 77.3%, 54.5%, 81.1%, 63.6%, 77.3%,72.7%, 100% and 100% respectively as a result of PCR analyses [38]. In addition, they concluded that all of the integron positive strains were resistant to cefepime and ceftazidime. Budak et al. found that 7 of 14 beta-lactamase- and aminoglycoside resistant P. aeruginosa strains were found to be both OXA-2- and OXA-10-positive and PER-1 [39]. It was seen that 2 isolates gave a PCR product with a 5’-3’ class I integron PCR.
Integrons are natural expression vectors that permit the insertion of antibiotic resistance genes by a site-specific recombinational mechanism. It has been shown that the P. aeruginosa strains that have different RAPD-PCR patterns have different integrons. We detected that in the two strains that were group representative blaPER-1 have relationship with CLASS-1 integrons. It would be necessary to support this relationship with DNA sequence analyses. We performed that PER-1 enzyme is common in our hospital and their clonal diversity indicates horizontal dissemination, the association between blaPER-1 and CLASS-1 integrons might accelerate dissemination of this gene.
References
- Gül M, Sensoy A, Çetin B, Korkmaz F, Seber E. Investigation of susceptibility of nosocomial Pseudomonas aeruginosa strains against ceftazidime by E-test and disc diffusion method. Turk Mikrobiyol Cem Derg 2004; 34: 33-36.
- Livermore DM. beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev 1995; 8: 557-584.
- Luzzaro F, Mantengoli E, Perili M, Lombardi G, Orlandi V, Orsatti A. Dynamics of a nosocomial outbreak of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended spectrum ß- lactamase. J Clin Microbiol 2001; 39: 1865-1870.
- Pollack M. Pseudomonas aeruginosa, In Mandell GL, Bennett JE, and Dolin R(ed), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Edinburgh, Scotland. 2000; 2310-3235.
- Pagani L, Migliavacca R, Pallecchi L, Matti C, Giacobone E. Emerging extended-spectrum beta-lactamases in Proteus mirabilis. J Clin Microbiol 2002; 40: 1549-1552.
- Pagani L, Mantengoli E, Migliavacca R, Nucleo E, Pollini S, Spalla M. Multifocal Detection of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum ß lactamase in Northern Italy. J Clin Microbiol 2004; 42: 2523-2529.
- Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of a novel extended-spectrum ß-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 1993; 37: 962-969.
- Vahaboglu H, Ozturk R, Aygun G, Coskunkan F, Yaman A, Kaygusuz A. Widespread detection of PER-1-type extended-spectrum ß-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother 1997; 41: 2265-2269.
- Vahaboglu H, Hall LMC, Mulazimoglu L, Dodanli S, Yildirim I, Livermore DM. Resistance to extended-spectrum cephalosporins, caused by PER-1 ß-lactamase, in Salmonella typhimurium from Istanbul, Turkey. J Med Microbiol 1995; 43: 294-299.
- Weldhagen GF, Poirel L, Nordmann P. Ambler Class A extended-spectrum b-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob Agent Chemother 2003; 47: 2385-2392.
- Lee S, Park YJ, Kim M, Lee HK, Han K, Kang CS. Prevalence of Ambler class A and D b-lactamases among clinical isolates of Pseudomonas aeruginosa in Korea. J Antimicrob Chemother 2005; 56: 122-127.
- Endimiani A, Luzzaro F, Pini B, Amicosante G, Rossolini GM, Toniolo AQ. Pseudomonas aeruginosa bloodstream infections: risk factors and treatment outcome related to expression of the PER-1 extended-spectrum beta-lactamase. BMC Infect Dis 2006; 6: 52.
- Yan JJ, Tsai SH, Chuang CL, Wu JJ. OXA-type betalactamases among extended-spectrum cephalosporinresistant Pseudomonas aeruginosa isolates in a university hospital in southern Taiwan. J Microbiol Immunol Infect 2006; 39: 130-134.
- Aubert D, Poirel L, Chevalier J, Leotard S, Pages JM, Nordmann P. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2001; 45: 1615-1620.
- Eraç B, Gülay Z. Molecular epidemiology of PER-1 extended spectrum beta-lactamase among gram-negative bacteria isolated at a Tertiary Care Hospital. Folia Microbiol 2007; 52: 535-541.
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 1995; 15: 593-600.
- Cantón R, Coque TM, Baquero F. Multi-resistant Gram-negative bacilli: from epidemics to endemics. Curr Opin Infect Dis 2003; 16: 315-325.
- Iraj N, Azita T, Zinab F, Kobra A. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbial 2013; 5: 36-41.
- Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 1995; 39: 185-191.
- Ruiz-Martínez L, López-Jiménez L, Fusté E, Vinuesa T, Martínez JP. Class 1 integrons in environmental and clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents 2011; 38: 398-402.
- Peng CF, Lee MF, Fu HT. Characterization of class 1 integrons and antimicrobial resistance in CTX-M-3-producing Serratia marcescens isolates from southern Taiwan. Jpn J Infect Dis 2007; 60: 250-256.
- Penteado AP, Castanheira M, Pignatari AC. Dissemination of bla(IMP-1)-carrying integron In86 among Klebsiella pneumoniae isolates harboring a new trimethoprim resistance gene dfr23. Diagn Microbiol Infect Dis 2009; 63: 87-91.
- Gu B, Tong M, Zhao W, Liu G, Ning M, Pan S. Prevalence and characterization of Class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol 2007; 45: 241-243.
- Clinical and Laboratory Standarts Institue. Performance standards for antimicrobial susceptibility testing; Sixteenth Informational Supplement. CLSI document 2006; M100- S16. Pennsylvania.
- Jiang X, Zhang Z, Li M, Zhou D, Ruan F, Lu Y. Detection of extended-spectrum- lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2006; 50: 2990-2995.
- Yetkin G, Kuzucu C, Durmaz B, Durmaz R, Cizmeci Z, Iseri L. Molecular typing of methicillin-resistant Staphylococcus aureus isolated from bloodstream infections in a university hospital. Turk J Med Sci 2009; 39: 959-968.
- Poirel L, Menuteau O, Agoli N, Cattoen C, Nordmann P. Outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J Clin Microbiol 2003; 41: 3542-3547.
- Poirel L, Cabanne L, Vahaboglu H, Nordmann P. Genetic environment and expression of the extended-spectrum ß-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob Agents Chemother 2005; 49: 1708-1713.
- Danel F, Hall LM, Gur D, Akalin HE, Livermore DM. Transferable production of PER-1 beta-lactamase in Pseudomonas aeruginosa. J Antimicrob Chemother 1995; 35: 281-294.
- Aktas Z, Poirel L, Salcioglu M, Özcan PE, Midilli K, Bal Ç. PER-1 and OXA-10 like beta-lactamases in ceftazidime-resistant Pseudomonas aeruginosa isolates from intensive care unit patients in Istanbul, Turkey. Clin Microbiol Infect 2005; 11: 193-198.
- Günseren F, Mamikoglu L, Öztürk S, Yücesoy M, Biberoglu K, Yulug N. A surveillance study of antimicrobial resistance of gram-negative bacteria isolated from intensive care units in eight hospitals in Turkey. J Antimicrob Chemother 1999; 43: 373-378.
- Kolayli F, Gacar G, Karadenizli A, Saniç A, Vahaboglu H. PER-1 still widespread in Turkish hospitals among Pseudomonas aeruginosa and Acinetobacter spp. FEMS Microbiol Lett 2005; 249: 241-245.
- Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum b-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns 2010; 36: 70-74.
- Yong D, Shin JH, Kim S, Lim Y, Yum JH, Lee K. High prevalence of PER-1 extended-spectrum ß-lactamase producing Acinetobacter spp. in Korea. Antimicrob Agents Chemother 2003; 47: 1749-1751.
- Fonseca EL, Vieira VV, Cipriano R, Vicente ACP. Class-1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol Med Microbiol 2005; 44: 303-309.
- Severino P, Magalhaes VD. The role of integrons in the dissemination of antibiotic resistance among clinical isolates of Pseudomonas aeruginosa from an intensive care unit in Brazil. Res Microbiol 2002; 153: 221-226.
- Zhu Y, Yi Y, Yang X, Lv N, Li J. Discovery of new structure of class 1 integron in MDR Pseudomonas aeruginosa and its association with drug-resistance. Wei Sheng Wu Xue Bao 2013; 53: 927-932.
- Kouchaksaraei FM, Shahandashti EF, Molana Z, Kouchaksaraei MM, Asgharpour F, Mojtahedi A, Rajabnia R. Molecular Detection of integron genes and pattern of antibiotic resistance in Pseudomonas aeruginosa strains isolated from intensive care unit, Shahid Beheshti Hospital, north of Iran. 2012; 1: 4.
- Budak F, Kasap M, Kolalyli F, Karadenizli A, Vahaboglu MH. Integron-associated resistance genes among multidrug-resistant Pseudomonas aeruginosa isolated from clinical specimens. Turk J Med Sci 2012; 42: 149-156.