Case Report - Journal of Cell Science and Mutations (2017) Volume 1, Issue 1
Investigation of microbial community associated with mediastinitis by 16S ribosomal DNA sequencing
Ajay Kumar Mishra*, Pierre-Edouard FournierResearch Unit on Emerging Infectious and Tropical Diseases, Faculty of Medicine, Aix-Marseille University, France
- *Corresponding Author:
- Ajay Kumar Mishra
Research Unit on Emerging Infectious and Tropical Diseases
Faculty of Medicine, Aix-Marseille University
France
Tel: 33 4 91 39 65 00
E-mail: ajaymishra24@gmail.com
Accepted date: March 27, 2017
Citation: Mishran AK and Fournier PE. Investigation of microbial community associated with mediastinitis by 16S ribosomal DNA sequencing.. Adv Cel sci tissue cul 2017;1(1):1-4.
DOI: 10.35841/cell-science.1.1.9-12
Visit for more related articles at Journal of Cell Science and MutationsAbstract
Here, we document the case of mediastinitis in an adult individual, developed upon contiguous spread of oesophageal infection. Our findings are noteworthy, because the presence of Prevotella oris , P. veroralis , Dialister pneumosintes , Klebsiella pneumoniae in tissue sample from mediastinum cannot be identified by standard culturing and Gram-staining. Importantly, however, 16S rDNA sequencing did lead to an unambiguous diagnosis and guided successful antimicrobial therapy.
Introduction
The wide range of esophageal disorders includes infectious and inflammatory processes, trauma and perforation, and benign and malignant neoplasms. The most common complications include mediastinitis secondary to esophageal perforation or postoperative anastomotic leak, or both; empyema due to fistula formation; and aspiration pneumonia [1]. Mediastinitis refers to inflammation of the mediastinal structures that lie between the right and left pleura between the thoracic inlet and the diaphragm. Mediastinitis can be divided into the acute and chronic forms according to the etiology and the clinical course [2]. The main causes of acute mediastinitis include esophageal perforation, post-operative complications and the spread of an adjacent infection [1]. The incidence rate of mediastinitis is low, ranging from 0.2% to 5% among patients submitted to median sternotomy and acute mediastinitis mortality has been estimated to range from 15.4% to 50% [2,3]. Notable among the infectious agents found in cultures obtained from inflamed mediastinal tissue are Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas sp and Escherichia coli, the last having been correlated with a high mortality rate [4,5] Mixed gram-positive and gram-negative infections account for approximately 40% of cases. Isolated gram-negative infections are rare causes. However, in some cases, the exudate culture may be negative, which is usually due to previous use of antibiotics [5].
Amplification, cloning and sequencing of the conserved and variable regions of the 16S rRNA gene has been widely applied to characterize or detect bacterial diversity in diseased or healthy human tissue samples from various sites within the body. Over the past decade, 16S rDNA amplification and sequencing was successfully useful in the microbiological diagnosis of brain abscesses [5], infected prosthetic joints [6], meningitis [7], osteomyelitis [8], septic arthritis [9], spondylodiscitis [10] and mediastintis [11].
Here, we report the results of an investigation of the bacterial diversity and clinical aspects of acute mediastinitis by the analysis of 16S rDNA libraries, in an effort to contribute to the ongoing characterization of flora associated with.
Materials and Methods
Clinical samples
Potential case of pleuro-mediastinitis complicating diagnosed at the hospital Saint-Marguerite, Marseille, France identified via infection control files and medical record discharge code. The case met the following criteria of mediastinitis-infection of the region within the thorax between the pleural sacs that is bounded inferiorly by the diaphragm, superiorly by the aperture of thorax, anteriorly by the sternum and costal cartilages, and posteriorly by the thoracic vertebrae. Tissue samples were obtained intra operatively by biopsy from within the mediastinum.
DNA extraction and purification
Clinical samples were incubated for 1 h at 55°C with proteinase K and lysis buffer. DNA was then extracted using NucleoSpin Tissue isolation kit (CLONTECH Laboratories Inc., Palo Alto, Calif.) according to the manufacturer's instructions. The final elution volume was 50 μl and 2 μl of DNA extracts were used for each PCR analysis.
Precautions and PCR pre-treatment with DNase I
Precautions taken to avoid laboratory contamination included physical separation of the pre-PCR areas used for sample preparation, DNA extraction, and preparation of the reaction mixtures from the post-PCR areas. Biosafety hoods and positive-displacement pipettes or barrier tips were used during the handling of specimens and reagents to avoid carryover and amplicon contamination. PCR premix (Taq polymerase, buffer and dNTPs; QIAGEN S.A, France) and RNase free water from the DNA extraction kit, which was used for resuspension of extracted DNA, were treated with DNase I (DNase I Bovine pancreas type IV, D-5025, SIGMA) (0.09 and 0.025 units of enzyme per μl of premix and water, respectively), and incubated for 1 h at 37°C. After incubation, DNase I was heat inactivated (1 h at 84°C) and the premix was cooled to 4°C before addition of primers and sample DNA to perform the PCR reaction.
PCR amplification and cloning of polymicrobial 16S rDNA
Polymicrobial 16S rDNA from the clinical samples was amplified using bacterial universal 16S rDNA primers fD1 (5´- AGAGTTTGATCCTGGCTCAG-3´) and rD1 (5´- AAGGAGGT GATCCAGCC-3´) [12]. Each 25-μl PCR reaction consisted of 100 ng of ploymicrobial DNA, 100 mM of each deoxynucleotide triphosphate, 0.2 mM of each primer, 1 X Taq buffer (20 mM TRIS-HCl, 100 mM KCl, 15 mM MgCl2, 10 mM(NH4)2SO4, 0.5% Tween 20, Stabalizer; pH 9.0), 1 U of HotStar Taq DNA Polymerase (QIAGEN S.A, France). Amplifications were performed in an Eppendorf thermal cycler ((MJ Research, Waltham, Mass.). The PCR reaction was an initial step of 95°C for 15 min then 35 cycles of: 94°C for 30 s, 55°C for 1 min, 72°C for 2 min followed by a final extension of 72°C for 10 min. The PCR products were resolved on a 1.5% agarose gel containing 0.5 mg/mL ethidium bromide and visualised under UV light. Gel photographs were scanned through a Gel Doc System (Bio-Rad Laboratories, Inc., USA). The single amplified band was purified using a QIAquick PCR purification kit (QIAGEN S.A, France), ligated to a plasmid (pGEM-T Easy System Promega, Madison, WI, USA) and transformed in Escherichia coli DH5α cells, according to the manufacturer’s instructions. Recombinants were selected on the basis of white/blue colonies and plasmids were purified using a commercial kit (QIAprep®, QIAGEN S.A, France). Screening of recombinants was performed by 1% agarose-gel electrophoresis with ethidium bromide staining. Two negative controls were included in each PCR series: a sample preparation control with sterile water that went through the same DNA extraction steps as the clinical samples and a reagent control with sterile water as a template.
Sequencing and phylogenetic identification (BLAST analysis)
Seventy recombinant clones per library were selected randomly for 16S rDNA sequencing reactions, performed in a DNA thermal cycler. Sequencing reactions were performed in both the directions with fD1, rD1 and two internal primers 800F (5´-ATTAGATACCCTGGTAG-3´) 800R (5´- CTACCAGGGTATCTAAT-3´) using an ABI Prism 310 automated sequencer (Applied Biosystems, Foster City, CA, USA). The temperature profile included an initial step at 96°C for 2 min, followed by 30 cycles of 96°C for 30 s, 50°C for 30 s and 60°C for 4 min. Reactions were performed in 10 ml mixtures containing 250 ng template DNA, 1 μl Big Dye Terminator Ready version 3.0 (Applied Biosystems), 0.5 μM primer and 3 μl sequencing buffer (200 mM Tris-HCl, pH 9.0, 5 mM MgCl2).
Comparative sequence analyses and databases
Contiguous sequences were assembled with the ChromasPro (Version: 2.33) software package, followed by analysis with Chimera Check (RDP II; http://rdp.cme.msu.edu) to eliminate chimeric molecules. The assembled sequences were grouped into clusters (phylotypes) according to a 99% minimum similarity rule [13]. For phylogenetic identification, sequences were compared with sequences of known bacteria listed in official databases using the BLAST program available at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). One representative of each phylotype was selected and submitted to the BLASTN algorithm. The criteria used to assign a species name were 98-100% homology to more than one GenBank sequence of the same species [14].
Results and Discussion
Acute mediastinitis continues to be a much feared complication of various diseases and procedures. Before the development of modern cardiovascular surgery, cases of acute mediastinitis usually arose from either perforation of the esophagus or from contiguous spread of odontogenic or retropharyngeal infections. However, in modern practice, most cases of acute mediastinitis result from complications of cardiovascular or endoscopic surgical procedures [15]. In our case patient has no surgical history and has oesophageal ulceration so it is believed that patient developed bacterial mediastinitis from contiguous spread of oesophageal infection. In this study, exudate was collected into sterile tube using sterile devices and surgical procedures, virtually eliminating the risk of perioperative contamination of specimen. Culture of the exudate revealed that the acute mediastinitis was caused by multiple microbes. Enterococcus faecalis, Stenotrophomonas maltophilia, Streptococcus anginosus and Streptococcus species were detected by culture-based technique. Direct 16S rDNA PCR amplification and sequencing identified polymicrobial infections (Table 1).
Table 1. Genotypical identification of the bacterial communities found in the mediastinitis patient.
| Phylogenetic affiliation | % Query coverage | % Similarity | Accession number |
|---|---|---|---|
| Prevotella oris | 99 | 100 | KF135659 |
| Preotella veroralis | 99 | 100 | KF135660 |
| Dialister pneumosintes | 100 | 100 | KF135661 |
| Klebsiella pneumoniae | 100 | 100 | KF135662 |
| Streptococcus anginosus | 99 | 100 | KF135663 |
| Streptococcus intermedius | 100 | 100 | KF135664 |
| Enterococcus faecalis | 100 | 100 | KF135665 |
| uncultured gamma proteobacterium clone HLB-37 | 98 | 100 | JF795440 |
| uncultured gamma proteobacterium clone HLB-40 | 100 | 100 | KF135666 |
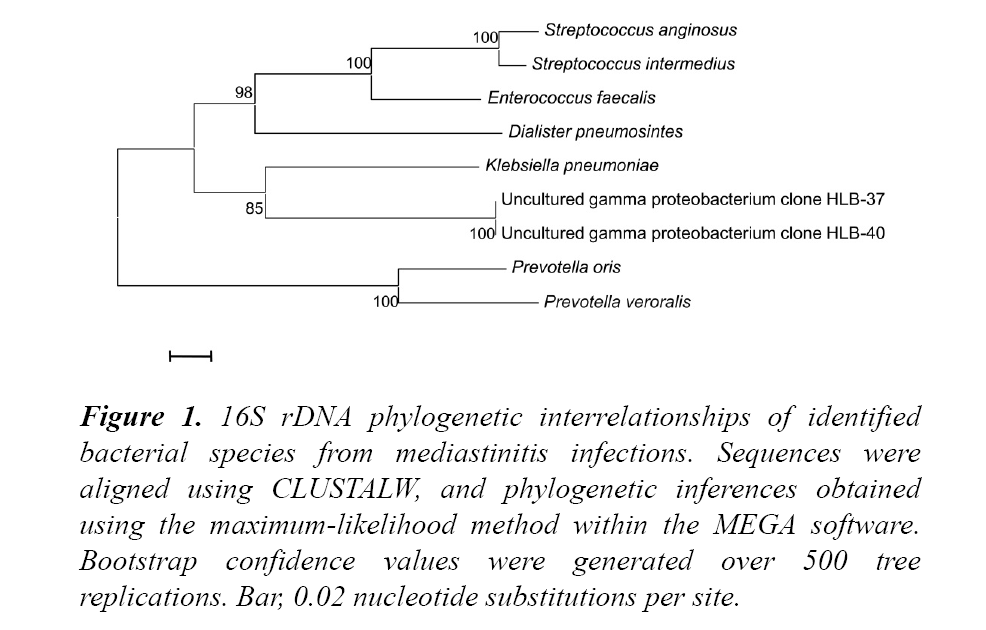
It identified Prevotella oris, P. veroralis, Dialister pneumosintes, Klebsiella pneumoniae, S. anginosus, S. intermedius, E. faecalis and two uncultured gamma proteobacterium according to the established 98% nucleotide identity threshold shown in Figure 1.
Figure 1: 16S rDNA phylogenetic interrelationships of identified bacterial species from mediastinitis infections. Sequences were aligned using CLUSTALW, and phylogenetic inferences obtained using the maximum-likelihood method within the MEGA software. Bootstrap confidence values were generated over 500 tree replications. Bar, 0.02 nucleotide substitutions per site.
This parameter is in accordance with a previously proposed species definition, based on DNA-DNA reassociation assays [14], and lies within the range of values employed by similar studies, which vary from 98% [16] to 99% [17]. Only S. maltophilia was only found by culture and was not detected by 16S rDNA gene sequencing. In both cases, other organisms S. anginous and E. faecalis were consistently found by culture and molecular detection. This study has demonstrated that the result obtained by direct PCR amplification and sequencing for bacterial identification was more sensitive and precise than phenotypic identification. After the result of the culture is known, the antibiotic therapy is adjusted according to the result obtained in the exam. Antibiotic therapy in combination with mediastinal drainage was used. In order to eliminate grampositive, gram-negative and anaerobic microorganisms broad spectrum antimicrobial combination (Tienam/Vancomycin/ Gentamicin) was administrated. Patient showed satisfactory improvement with persistence of fever at 38.5°C. However, the effusion of empyema from right pleura showed the presence S. anginous and E. faecalis by both culture as well as 16S rRNA PCR methods. Thoracotomy was performed in this case to drain the mediastinal and pleural effusions. Samples taken after surgery were negative for bacteremia by both culture and PCR methods. This report also indicates that a precise and rapid identification of unusual bacteria species by use of 16S rDNA based amplification, cloning and sequencing.
References
- Giménez A, Franquet T, Erasmus JJ, et al. Thoracic complications of esophageal disorders. Radiographics. 2002;22:S247-S258.
- Macrí P, Jiménez MF, Novoa N, et al. Descriptive of a series of patients diagnosed with acute mediastinitis. Arch Bronconeumol 2003;39:428-430.
- Konvalinka A, Errett L, Fong IW. Impact of treating Staphylococcus aureus nasal carriers on wound infections in cardiac surgery. J Hosp Infect 2006;64:162-168.
- Sancho LM, Minamoto H, Fernandez A, et al. Descending necrotizing mediastinitis: a retrospective surgical experience. Eur J Cardiothorac Surg. 1999;16:200-205.
- Al Masalma M, Raoult D, Roux V. Phocaeicola abscessus gen. nov., sp. nov., an anaerobic bacterium isolated from a human brain abscess sample. Int J Syst Evol Microbiol. 2009;59:2232-2237.
- Vandercam B, Jeumont S, Cornu O et al. Amplification-based DNA analysis in the diagnosis of prosthetic joint infection. J Mol Diagn. 2008;10:537-543.
- Arosio M, Nozza F, Rizzi M, et al. Evaluation of the MicroSeq 500 16S rDNA-based gene sequencing for the diagnosis of culture-negative bacterial meningitis. New Microbiol. 2008;31:343-349.
- Welkerling H, Geissdorfer W, Aigner T, Forst R. Osteomyelitis of the ulna caused by Porphyromonas gingivalis. J Clin Microbiol. 2006;44:3835-3837.
- Rosey AL, Abachin E, Quesnes G, et al. Development of a broad-range 16S rDNA real-time PCR for the diagnosis of septic arthritis in children. J Microbiol Methods. 2007;68:88-93.
- Fuursted KM, Arpi M, Lindblad BE, et al. Broad-range PCR as a supplement to culture for detection of bacterial pathogens in patients with a clinically diagnosed spinal infection. Scand J Infect Dis. 2008;40:772-777.
- Martínez J, Martorell J, Abarca ML, et al. Pyogranulomatous pleuropneumonia and mediastinitis in ferrets (Mustela putorius furo) associated with Pseudomonas luteola infection. J Comp Pathol. 2012;146:4-10.
- Weisburg WG, Barns SM, Pelletier DA, et al. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697-703.
- Hutter G, Schlagenhauf U, Valenza G, et al. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiol. 2003;149:67-75
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152-5.
- Katabathina VS, Restrepo CS, Martinez-Jimenez S, et al. Nonvascular, nontraumatic mediastinal emergencies in adults: a comprehensive review of imaging findings. RadioGraphics. 2011;31:1141-1160.
- Munson MA, Pitt-Ford T, Chong B, et al. Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res. 2002;81:761-766.
- Hutter G, Schlagenhauf U, Valenza G, et al. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiol. 2003;149:67-75.