Research Article - Journal of Systems Biology & Proteome Research (2018) Journal of Systems Biology & Proteome Research (Special Issue -2018)
Insights into interaction profile and inhibitory potential of amentoflavone with ?-glucosidase, tyrosinase and 15-lipoxygenase as validated therapeutic targets.
Tomisin Happy Ogunwa1,2*1Centre for Bio-computing and Drug Development, Adekunle Ajasin University, Akungba-Akoko, Ondo State, Nigeria.
2Department of Biochemistry, Adekunle Ajasin University, Akungba-Akoko, Ondo State, Nigeria.
- Corresponding Author:
- Tomisin Happy Ogunwa
Centre for Bio-computing and Drug Development
Adekunle Ajasin University
Akoko, Ondo State, Nigeria
Accepted Date: May 03, 2018
Citation: Ogunwa TH. Insights into interaction profile and inhibitory potential of amentoflavone with α-glucosidase, tyrosinase and 15-lipoxygenase as validated therapeutic targets. J Syst Biol Proteome Res. 2018;2(1):10-20
Abstract
Therapeutic benefits of amentoflavone, a 5',8"-biapigenin isolated from numerous medicinal plants, in the management of human ailments has been reported. However, the mechanisms of action and precise interaction of this biflavonoid with protein targets for such bioactivities have not been exhaustively documented. The present study shows the first binding model of amentoflavone with few selected human proteins (α-glucosidase, tyrosinase and 15-lipoxygenase) as validated therapeutic targets using computational procedures. Results obtained divulged amentoflavone interference with the metal ions within the active site of tyrosinase and lipoxygenase as part of its inhibitory mechanism against the proteins. The predicted inhibitory potential of the biflavonoid against 15-lipoxygenase correlates with IC50 value (0.04 µM). Its binding poses were comparable to that of acarbose on α-glucosidase with highest affinity (-11.4 kcal/mol) while the modest affinity (-9.4 kcal/mol) for tyrosinase indicates moderate inhibition for the compound possibly due to its interaction with only one of the two copper ions. It was observed that amentoflavone stably occupied the active sites of all the proteins with lesser ?G values and engaged one of its monoflavonoid subunits for penetration into binding pocket, preventing substrate access, binding and conversion. However, the relatively bulky structure of amentoflavone was prone to steric hindrance within the binding pockets. The polyhyroxyl groups of the biflavonoid participate critically in the formation of hydrophilic interaction. The aromatic rings enjoyed hydrophobic interactions with residues side chain and π-π stacking with the phenolic rings of aromatic amino acids. The unique binding configurations and interactions obtained against each protein in this study may be helpful in explaining the subtle differences of the biflavonoid potency (IC50 values). The data also validate amentoflavone as a useful candidate in treatment of diabetes, inflammation and hyperpigmentary disorders.
Keywords
Amentoflavone, α-glucosidase, Biflavonoid, 15-Lipoxygenase, Binding model, Tyrosinase.
Introduction
Amentoflavone is one of the plant-derived biflavonoids currently being explored for potent pharmacological activities. It is considered as the dimeric form of monoflavonoid apigenin and has been identified in various medicinal plants with known bioactivities [1,2]. Biflavonoids are unique biologically active naturally-occurring compounds for which the attentions of researchers have been drawn over the years. They have been reported to possess higher potency and effectiveness compared to their corresponding monoflavonoid components in various experiments [3]. A biflavonoid can be linked by C-C or C-O-C bonds in homo or hetero subunits combination. Flavone-flavone, flavanone-flavone and flavanone-flavanone are the common subunits found in plant-derived biflavonoids [1,4]. However, the connecting linkages are present at diverse positions yielding more than a hundred biflavonoids which have been isolated and structurally characterized till date [1,5]. Recent reports have shown that synthetic biflavonoids are now produced for specific therapeutic uses [6]. Biflavonoids are generally known for a wide number of pharmacological potentials in different countries and they are found in limited number of plants with interesting medicinal and pharmacological properties. For instance, amentoflavone was firstly identified in the Selaginella species (Selaginella pachystachys, Selaginella tamariscina (Beauv.) Spring and Selaginella nipponica) in 1971 by Okigawa et al. [7] and to date, more than 120 locally available plants with folkloric medicinal applications have been named across the world to contain this biflavonoid [8]. Such important plants include Garcinia livingstonei, Ginkgo biloba, Selaginella tamariscina, Ginkgo subelliptica, Hypericum perforatum, Xerophyta plicata, Chamaecyparis obtusa, Ouratea multiflora, Juniperus phoenica, Semecarous anacardium, Selaginella sellowii, Ochna schweinfurthiana, Selaginella doederleinii, Ouratea stipulata and, Euphorbiaceae, Cupressuceae spp, Calophyllaceae, Podocarpaceae plant families [2,7].
As common to polyphenolic compounds, potent pharmacological activities have been documented for amentoflavone by numerous researchers. While Coulerie et al. [9] reported the anti-viral effect of amentoflavone, Ndongo and coworkers [10] described the cytotoxic effect of the biflavonoid on various cancer cell lines.
The apigenin dimer possesses anti-diabetic, hepatoprotective, antimalarial, antiinflammatory, antihyperlipidemic and antioxidation activities [2,10-15]. Amentoflavone has also shown beneficial therapeutic potentials on central nervous systems (neuroprotection) and cardiovascular system [16]. The biflavonoid was reported for its potent in vitro antimicrobial (antibacterial and antifungal) effects against diverse pathogenic fungal strains [17]. Other researchers have also observed the antidepressant, radioprotective, anti-senescence, antiangiogenic as well as anti-ulcerative colitis properties of the naturally-occurring biflavonoid [18-23].
In frantic efforts to identify the target for the known bioactivities of amentoflavone, various in vitro and in vivo experiments have been carried out in recent time. Among the reported human proteins (enzymes and receptors) susceptible to amentoflavone antagonistic effect are phosphodiesterases (PDE3 and PDE5) which are found in adipose tissue and other tissues [8,24,25]. The biflavonoid also displayed inhibitory activity against nitric oxide synthase, fatty acid synthase, acetylcholinesterase and cytochrome proteins (CYP3A4 and CYP2C9) [13,26,27]. Using both in vitro and in silico experiments, Lee et al. [1] documented the allosteric inhibitory potential of amentoflavone against protein tyrosine phosphatase 1B (PTP1B). The biflavonoid inactivated NF-kß [28] and modulated benzodiazepine GABA (A) receptor at the allosteric site [29]. Hanrahan and coworkers [29] also reported the biflavonoid for its antagonistic mechanism on kopioid receptor. It can also inhibit the enzymatic function of vascular endothelial growth factors (VEGFs), human cathepsin B (1) and α-amylase [30-32].
Human 15-Lipoxygenase, tyrosinase and α-glucosidase are few of the recently identified targets of amentoflavone for the anti-inflammatory, anti-browning and anti-hyperglycemic activities respectively. However, the mechanism of interaction between the biflavonoid and these proteins has not been clearly unraveled. Previously, our research team has reported the interaction patterns of amentoflavone against α-amylase [33], as well as morelloflavone with few other Garcinia biflavonoids against 3-hydroxy-3-methylglutaryl-CoA (HMG-COA) reductase [34]. As part of these ongoing studies to wholly investigate the mechanisms responsible for beneficial health effects of naturally-occurring biflavonoids, computational approach was adopted herein to build plausible binding models of amentoflavone with human α-glucosidase, 15-lipoxygenase and tyrosinase to provide insights into their precise interaction signatures towards understanding its mode of inhibition and potency against these proteins.
Materials and Method
Selection and preparation of target proteins
The crystal structure of proteins used in the current research was obtained from the Brookhaven protein data bank (http:// www.rcsb.org/pdb). Human α-glucosidase (maltase) (PDB ID: 3TOP), 15-lipoxygenase (PDB ID: 3V99) and tyrosinase (PDB ID: 2Y9X) were used as the starting coordinates. Each protein structure was visualized using the molecular graphics program PyMol intended for the structural visualization of proteins. The macromolecules were found in complex with ligand and crystallographic water molecules which were deleted prior molecular docking procedure. The active site of each protein was identified with reference to the co-crystallized ligand.
Preparation of reference ligands
Acarbose, linoleic acid mimetic and tropolone (the co-crystallized inhibitors) were used as the reference ligand for α-glucosidase, 15-lipoxygenase and tyrosinase respectively. The structure of acarbose (CID 41774) and tropolone (CID 10789) were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih. gov). The 3D format (sdf) of pyrimido[4,5-b][1,4]benzothiazine derivative previously reported as 15-lipoxygenase was prepared using ChemAxon software (http://www.chemaxon.com).
Preparation and optimization of the biflavonoid structure
The chemical structure of the biflavonoid (amentoflavone) used in this docking study was retrieved from NCBI PubChem database (http://www.ncbi.nlm.nih.gov/pccompound) with CID: 5281600 and prepared using ChemAxon software (https://www. chemaxon.com). Identification and selection of the biflavonoid were based on information from the literature. The other ligands were used as control ligands, one control ligand for each protein. Marvin-Sketch v15.11.30 was used to sketch the 2D-coordinates of the ligands. The structures were then cleaned up in 2D and converted to 3D geometry using the Conformers suit of the software based on the Merck molecular force field (MMFF94). The MDL SDfile (.sdf) format of the ligands was finally docked into the active site of the targets using the AutoDock Vina.
Validation of molecular docking procedures
A key strategy towards validation of docking procedure is to accurately regenerate both the binding pose and molecular interaction of the co-crystallized ligand on the X-ray structure of the protein structure. In this study, the ligand found at the binding site of each experimentally determined protein crystals was deleted. Then, each ligand (.sdf format) was separately prepared using Marvin sketch as described above and re-docked into the active site. The binding poses and molecular interaction, majorly hydrogen bond in this case, were compared to that of the x-ray diffraction crystal structures.
Molecular docking and scoring
For ligand docking and target-ligand complex analysis, Autodock Vina suite on PYMOL [35,36] was used. First, based on the already present co-crystallized ligand in the pdb file, the inhibitor binding site was defined with grid parameters and coordinate of origin (x, y and z) set as shown in Tables 1 and 2 to include all the amino acid residues at the active site. This gives enough space to enhance adequate ligand rotation and translation. The spacing between grid points was maintained at 0.375 angstroms. All optimized ligands were docked to the active site of the proteins. Throughout this in silico investigation, the rotatable bonds of the ligands were set to be free and a total of 10 docking runs were performed for each ligand with the number of modes set to 10 so as to achieve more accurate and reliable results. The best results obtained based on the binding configuration and binding affinity was chosen for further analysis.
| Protein name | Receptor (PDB ID) | Centre X (Å) | Centre Y (Å) | Centre Z (Å) |
|---|---|---|---|---|
| α-Glucosidase | 3TOP | -30.62 | 35.65 | 26.44 |
| Tyrosinase | 2Y9X | -10.02 | -28.82 | -43.6 |
| 15-Lipoxygenase | 3V99 | 8.48 | -79.18 | -25.58 |
Table 1. Grid center coordinates (X, Y and Z) used for the docking runs.
| Protein name | Crystal resolution (Å) | Spacing (Å) | X point (Å) | Y point (Å) | Z point (Å) |
|---|---|---|---|---|---|
| α-Glucosidase | 2.881 | 0.375 | 120 | 120 | 120 |
| Tyrosinase | 2.78 | 0.375 | 100 | 100 | 100 |
| 15-Lipoxygenase | 2.252 | 0.375 | 200 | 200 | 230 |
Table 2. Grid box spacing and parameters (X, Y and Z points) used in this study for the docking.
Data analysis
Protein-ligand complex visualization, interaction depiction and snapshots were achieved using PYMOL. Validation of the molecular interaction was done using Ligplot and Proteinsplus (an online server), especially for hydrophobic bond and piestacking interactions [37,38].
Results and Discussion
Herein, in silico experimental procedures were employed to prepare the binding models of amentoflavone with selected human therapeutic proteins including α-glucosidase, 15-lipoxygenase and tyrosinase to gain insights into the biflavonoid binding signature and molecular interaction as inhibitor of the proteins. The choice of the current study was informed by the dearth of information on the precise amentoflavone-targets interaction profile in the literature. The chemical structure of amentoflavone and its monomeric flavonoid (apigenin) are presented in Figure 1. Amentoflavone belongs to the biflavonoid class of naturallyoccurring biflavonoids and has attracted attention of researchers over the last few decades due to theirits pharmacological activities. It comprises of two apigenin monomers connected by a covalent C3' - C8" linkage [2,8]. It is known that the type of linkage present in biflavonoids is a key determining factor for them to gain particular molecular interaction pattern which can inhibit a protein as a target. The different positions of the hydroxyl and methoxyl moieties on the biflavonoids also influence their binding pose, fitness as well as interaction within a putative binding pocket of an enzyme and can eventually determine their inhibitory potential [1,33]. In a related manner, apigenin is an important flavonoid with numerous reported bioactivities in in vitro, in vivo and in silico experiments [39]. The potency of amentoflavone has been proven higher compared to apigenin in various reports [40-42]. Amentoflavone has been reported for its antidiabetic, anti-hyperglycemic, anti-browning, antitumor and anti-inflammatory properties in vitro [32,43,44]. Vigorous computational analyses are often used in the current trend of searching for possible targets for these bioactivities as well as inhibitors from natural and synthetic sources as potential replacements for the present day drugs in the management of diseases associated with the targets functions. It is thus envisaged that an in silico study, as carried out here, is crucial in providing corroborating information and revealing the specific interactions which make such bioactivities possible as well as the precise pattern of such interactions. As shown in Table 3, the calculated physicochemical properties substantiate amentoflavone and apigenin as biologically active natural compounds.
| Ligand | Chemical formular | Molecular weight | No of HB acceptor | No of HB donor | No of rotatable bonds |
|---|---|---|---|---|---|
| Amentoflavone | C30H18O10 | 538.464 | 10 | 6 | 3 |
| Apigenin | C15H10O5 | 270.24 | 5 | 3 | 1 |
Table 3. Physicochemical properties of the amentoflavone, its monoflavonoid.
Interaction profile of amentoflavone with humanα-glucosidase (maltase)
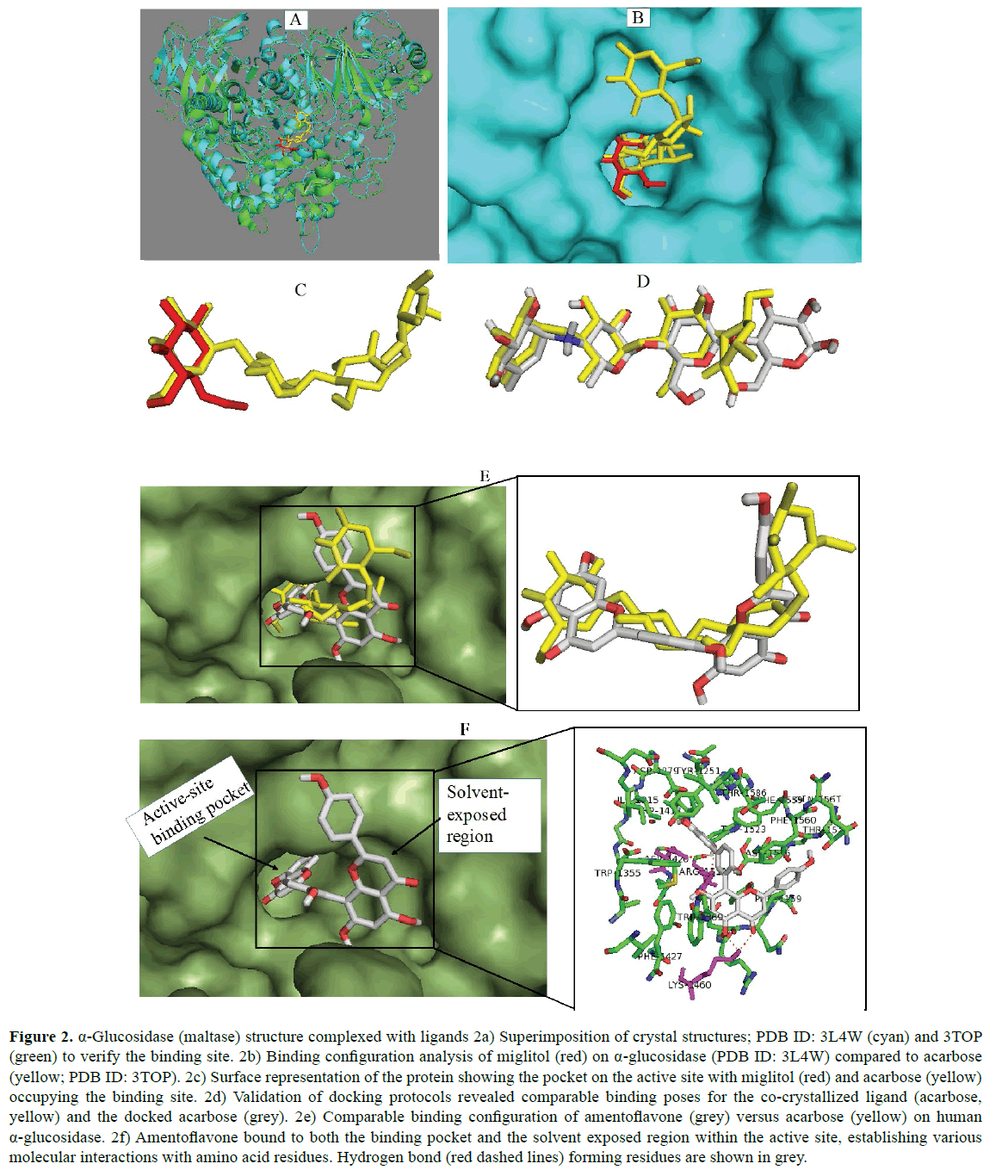
The protein α-glucosidase remains as one of the validated therapeutic targets in the management of diabetes mellitus especially in type 2 diabetes. Thus, the protein is a major molecular target for anti-diabetes drugs like acarbose, the control ligand in this study [45]. In order to have insights on the antidiabetic mode of action of amentoflavone, molecular docking study was performed against human α-glucosidase and the results obtained were analyzed. These are shown in Figure 2. First, two crystal structures of the enzyme co-crystallized with different inhibitors (acarbose and miglitol) were retrieved and superimposed (Figure 2a) to identify their binding location on the protein. Miglitol and Acarbose belong to the group of inhibitors currently deployed to manage blood glucose levels in type 2 diabetes, which targets digestive α-glucosidases as well as α-amylases for inhibition [46]. Analysis of their binding profile showed that they bind at the active site, buried deep into the active site. Acarbose displayed a much more occupation of the active pocket and the solvent exposed region (Figure 2c). The acarbose structure was therefore used to validate the reliability of the docking results. As revealed in Figure 2d, the in silico procedures regenerated a comparable binding pose for the reference ligand (acarbose) which suggests that the protocol used in this study could be relied upon to predict accurate binding mode of the compound evaluated. From the results, amentoflavone interacts directly with the maltase enzyme at the active site in a manner similar to acarbose Figure (2e) and can hence, prevent substrate access. The apigenin dimer can bind tightly with the active site of human maltase with favourable lesser ΔG value Figure (2f). This finding is in agreement with the reports of Aminudina et al. Compared to the control ligand (acarbose) with binding energy -8.8 kcal/mol, the binding energy (-11.4 kcal/mol) estimated for the biflavonoid against the enzyme (Table 4) indicates its very high affinity to the protein at the active site and thus, amentoflavone possesses potent inhibitory capacity against the biological functions of α-glucosidase. High affinity binding is probably due to its ability to fill up the binding pocket. Furthermore, the significantly higher affinity of the biflavonoid is also attributable to the increasing number of interaction of the subunits with amino acid residues in both the putative binding pocket and the solvent exposed area of the protein. Serendipitously, this result is consistent with 98.8% inhibitory activity and IC50 values of 6.4 μM and 8.09 μM reported for the biflavonoid in previous in vitro experiments [32,43]. Among the biological effects known for amentoflavone are hypoglycemic and antidiabetic effects. The current study has shown interaction with α-glucosidase as, at least a part, in the mechanisms for such bioactivities. Moreover, the results validate the biflavonoid as one of the antidiabetic ingredients in the medicinal plants containing it. Earlier, Kim et al. [47] has reported that amentoflavone has the potential to block the catalytic activity of yeast α-glucosidase. The molecular interaction analysis of the docking results herein indicated that amentoflavone forms 3 hydrogen bonds with residues Asp1420, Lys1460 and Arg1510 of the enzyme. In addition, the interaction results suggested π-π stacking between aromatic rings of the biflavonoid with that of the residue Phe1560 and Phe1559. Furthermore, the bulky structure of amentoflavone participated in hydrophobic interaction with Phe1559, Met1421 and Phe1560. These interactions contribute to the enzymeamentoflavone complex stability and are essential for inhibition of the protein catalytic functions by the apigenin dimer. In comparison, acarbose established complex hydrogen bonds with the enzyme. However, it lacks π-π stacking and hydrophobic interactions. This observation might have contributed to its reduced potency (9.95 μM) as previously reported [48] which is consistent with the affinity predicted for the compound in this study.
Figure 2: α-Glucosidase (maltase) structure complexed with ligands 2a) Superimposition of crystal structures; PDB ID: 3L4W (cyan) and 3TOP (green) to verify the binding site. 2b) Binding configuration analysis of miglitol (red) on α-glucosidase (PDB ID: 3L4W) compared to acarbose (yellow; PDB ID: 3TOP). 2c) Surface representation of the protein showing the pocket on the active site with miglitol (red) and acarbose (yellow) occupying the binding site. 2d) Validation of docking protocols revealed comparable binding poses for the co-crystallized ligand (acarbose, yellow) and the docked acarbose (grey). 2e) Comparable binding configuration of amentoflavone (grey) versus acarbose (yellow) on human α-glucosidase. 2f) Amentoflavone bound to both the binding pocket and the solvent exposed region within the active site, establishing various molecular interactions with amino acid residues. Hydrogen bond (red dashed lines) forming residues are shown in grey.
| Ligand | Protein (Target) | Estimated binding energy (kcal/mol) | No of hydrogen bond | Residues involved in hydrogen bond formation | Residues involved in hydrophobic interaction | Residues involved in π-cation stacking |
|---|---|---|---|---|---|---|
| Amentoflavone | α-Glucosidase | -11.4 | 3 | Asp1420, Lys1460, Arg1510 | Met1421, Phe1559, Phe1560 | Phe1560, Phe1559 |
| Tyrosinase | -9.4 | 2 | Asn260, His85 | Val283 | - | |
| 15-Lipoxygenase | -10.8 | 3 | Gln413, Gln363, Gln609 | Asn180, Leu607, Phe177, Ala410 | Phe177 |
Table 4. Estimated binding energy and interaction profile of amentoflavone with the proteins.
Interaction profile of amentoflavone with tyrosinase
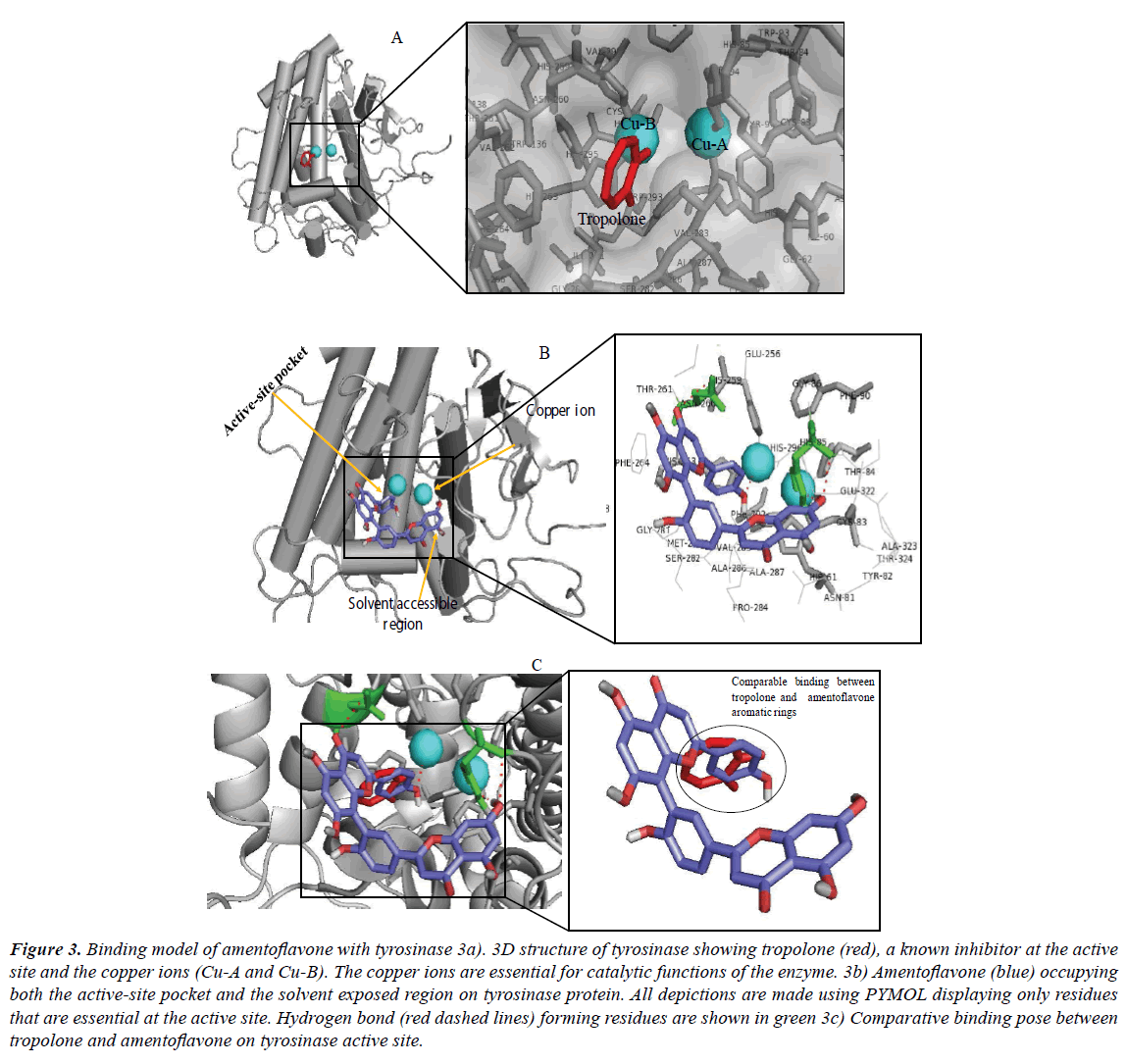
Inhibition of tyrosinase has evolved as a promising therapeutic concept in the treatment of human hyperpigmentary disorders including lentigo, freckles, senile lentigines, melasma, age spot and ephelide in human skin and, in agricultural uses (antibrowning in farm products). In addition, excessive production of melanin precursors often leads to generation of highly reactive compounds such as quinones metabolites which have been implicated in the initiation of cell damage or death [49-51]. Therefore, efforts have been put into the search for potent inhibitors of tyrosinase most importantly from natural products possibly due to their reduced toxicity compared to the synthetic agents. Amentoflavone, a plant-derived biflavonoid, was investigated as potential inhibitor of this enzyme here using computational approach. The docked result with the lowest binding energy and best binding pose was selected as presented in Figure 3. The crystal structure chosen as the starting coordinate of the protein (PDB ID: 2Y9X), as downloaded from protein databank, was found in complex with a co-crystallized inhibitor (tropolone). As seen in Figure 3a, the protein structure contains two cupper ions. Tyrosinase, the enzyme catalyzing the main and rate limiting steps in melanogenesis, has been described as a copper-containing metalloenzyme which is multifunctional exhibiting both monophenolase and diphenolase activity during mammalian melanin production [52,53]. Proper coordination of the copper ions is crucial for the enzymatic activity [54]. After docking, the observed binding free energy of amentoflavone was -9.4 kcal/mol. The result suggests a direct interaction between the biflavonoid and tyrosinase (Figures 3b and 3c), while the reduced affinity compared to α-glucosidase binding indicates that the compound moderately inhibited the activity of the enzyme (Table 4). However, amentoflavone displayed a better affinity relative to tropolone against tyrosinase (Table 5). The OH moieties on the aromatic rings of the biflavonoid formed 2 hydrophilic interactions with amino acid residues Asn260 and His85. Hydrophobic interaction was observed with Val283 while cation-π interaction was found with residue His263. These residues as well as their specific interactions have been significant in the inhibition of tyrosinase by known inhibitors. The cation-π interaction of His263 is also present in interaction with tropolone [54-56]. Much more importantly, amentoflavone interfered via hydrogen bond formation with one of two cupper molecules. This forms a part of the mechanism for tyrosinase inhibition by the biflavonoid. However, this interaction with only one of the copper ions (Cu-B) instead of both ions suggests the moderate activity of amentoflavone against the protein.
Figure 3: Binding model of amentoflavone with tyrosinase 3a). 3D structure of tyrosinase showing tropolone (red), a known inhibitor at the active site and the copper ions (Cu-A and Cu-B). The copper ions are essential for catalytic functions of the enzyme. 3b) Amentoflavone (blue) occupying both the active-site pocket and the solvent exposed region on tyrosinase protein. All depictions are made using PYMOL displaying only residues that are essential at the active site. Hydrogen bond (red dashed lines) forming residues are shown in green 3c) Comparative binding pose between tropolone and amentoflavone on tyrosinase active site.
This observation is also correlated by the reduced affinity of the compound (-9.4 kcal/mol). Potent inhibitors of tyrosinase possibly interact with the two copper ions. Within the enzyme active site, the copper molecule (Cu-B) is coordinated by four histidine residues. Specific synthetic or natural inhibitors interactions could displace the Cu2+-coordination or chelate the ions which are required for catalysis [49,57,58]. For instance, tropolone, which is reportedly one of the most effective inhibitors of tyrosinase was observed competing against substrates for binding to the copper ions at the active site of tyrosinase (Table 5). The potency of tropolone has been associated with chelation of the cupper ions and thus making them unavailable for the enzyme use. In this study, hydrogen bonding with copper B (Cu-B), hydrophobic interaction with residues Ser282, Phe292, Met280, Phe264, His259, Val283, His263, Ala286, His61 and Gly281 as well as cation-π interaction with residue His263 contribute to the tyrosinase-tropolone complex stability with IC50 4 μM [56]. The amino acid His65 is one of the residues responsible for the coordination of the Cu2+ ions via hydrophilic interaction. By establishing hydrogen bond with this amino acid residue, amentoflavone could distort the participation of His65 in the coordination and hence, eventually lead to inhibition of the protein catalytic function. Tropolone is reportedly the most potent synthetic inhibitor of tyrosinase which can effectively chelate the important copper ions within the active site (Figure 3a) [58,59]. A detailed observation of the amentoflavonetyrosinase complex revealed that the compound inserted the ring C of its second apigenin subunit into the binding pocket in a comparable manner to tropolone (Figure 3c).
| Control ligand | Protein | Estimated binding energy | No of hydrogen bond | Residues involved in hydrogen bond formation | Residues involved in hydrophobic interaction |
|---|---|---|---|---|---|
| (Target) | (kcal/mol) | ||||
| Acarbose | α-Glucosidase | -8.8 | 6 | His1580, Asp1279, Asp1526, Arg1510, Asp1157, Lys1460 | - |
| Tropolone | Tyrosinase | -6.4 | 1 | Cu-B | Phe264, Phe292, Gly281, His259, Val283, Ser282, His61, His263, Ala286 and Met280, |
| “Inhibitor 4d” | 15-Lipoxygenase | -7.5 | 1 | Ala672 | Ile406, Phe610, Phe555 |
Table 5. Estimated binding energy and interaction profile of the control ligands.
The other subunit conveniently covered the entrance to the active site to prevent substrate binding. Earlier, this biflavonoid has been reported as one of the inhibitors of tyrosinase from natural sources [44,59]. Inhibition of tyrosinase by amentoflavone may not be unconnected to the resorcinol component in the structure. It is widely known that this moiety is critical for enzyme inhibition. Not only that, a derivative of the biflavonoid (2,3-dihydro-4',4'"-di-O-methylamentoflavone) also showed anti-tyrosinase effect with IC50 98 μM in vitro [60]. It is envisaged that optimization of the amentoflavone scaffold might yield a useful agent in the treatment of melanin-related diseases.
Interaction profile of amentoflavone with human 15-lipoxygenase
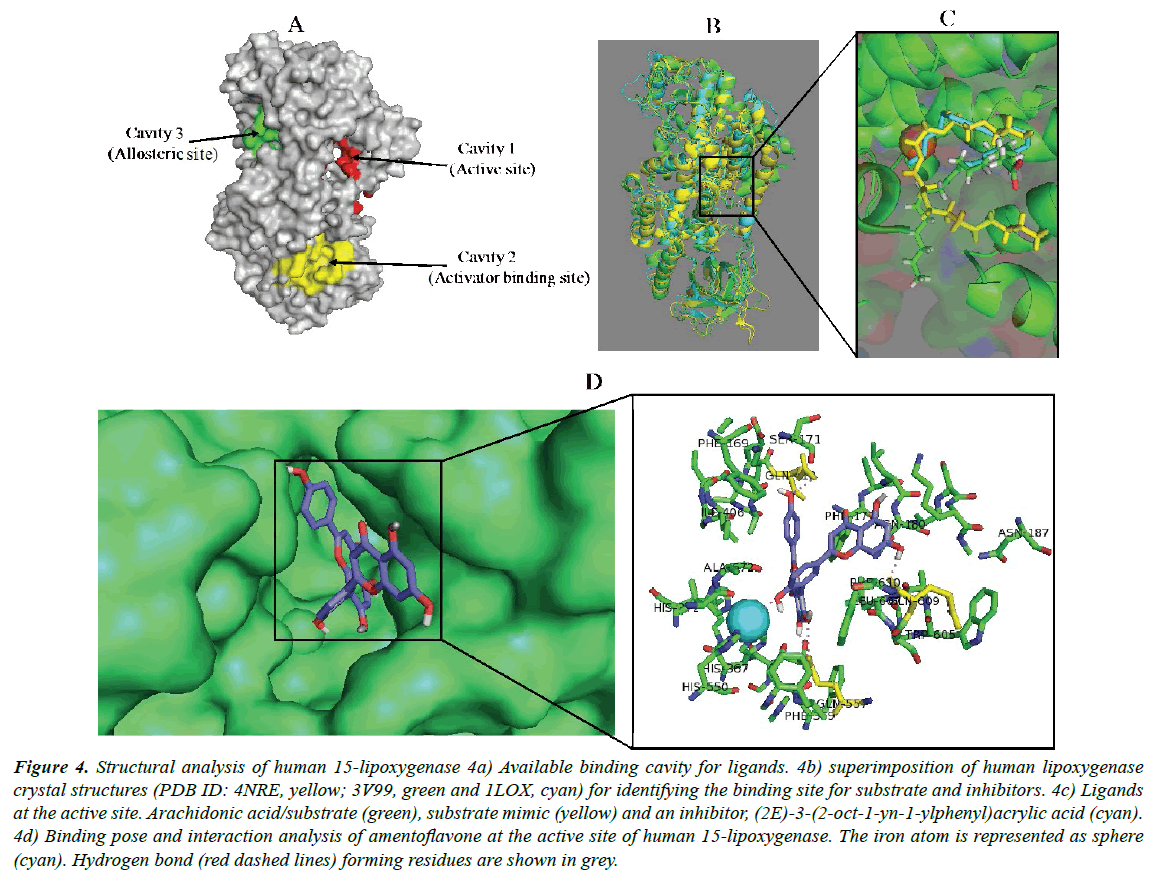
The structural analysis of the human 15-lipoxygenase showed that the protein has several binding cavities where ligands can interact (Figure 4a). Cavity 1 is the substrate binding pocket where competitive inhibitors also bind. Cavity 2 has been reported as an allosteric binding pocket for activators/effectors binding. Interaction of a ligand at this site induces structural change in the protein structure [61]. The possible role of cavity 3 has not been clearly defined till date. Identification of the binding pocket suitable for a ligand can determine the particular activity of an interacting ligand, either inhibition, activation or modulation. In the current study, the first step was to identify the binding location for amentoflavone on human 15-lipoxygenase. Blind docking was carried out using the coordinates shown in Table 1 and Table 2. By choosing a box size that completely covers the protein structure for molecular docking experiment, the ligand is allowed to search for its most suitable binding site on the protein. According to this study, amentoflavone relatively prefers the active site (Figure 4b) and binds in a similar manner to a co-crystallized competitive inhibitor, (2E)-3-(2-oct-1-yn- 1-ylphenyl) acrylic acid (Figure 4c). This indicates that the biflavonoid is an inhibitor of the protein displaying competitive inhibition type. The 15-lipoxygenase has been implicated in many human diseases since the enzyme participates in the metabolism of polyunsaturated fatty acids by catalyzing their oxidation to a variety of eicosanoids, which serve as secondary signal transducers with major impacts on human homeostasis [62]. The protein is involved in diseases including inflammatory responses, obesity, neurodegenerative disorders, cardiovascular disease, cancers and kidney diseases, and metabolic syndrome [63]. Therefore, inhibitors of this protein are sought as suitable agents for the necessary therapeutic applications including chemotherapy [64]. Amentoflavone has been identified in the leaves and whole plant of numerous medicinal plants and may account at least in part for their pharmacological properties including anticancer, antiobesity and antiinflammatory activities [2,8,64].
Figure 4: Structural analysis of human 15-lipoxygenase 4a) Available binding cavity for ligands. 4b) superimposition of human lipoxygenase crystal structures (PDB ID: 4NRE, yellow; 3V99, green and 1LOX, cyan) for identifying the binding site for substrate and inhibitors. 4c) Ligands at the active site. Arachidonic acid/substrate (green), substrate mimic (yellow) and an inhibitor, (2E)-3-(2-oct-1-yn-1-ylphenyl)acrylic acid (cyan). 4d) Binding pose and interaction analysis of amentoflavone at the active site of human 15-lipoxygenase. The iron atom is represented as sphere (cyan). Hydrogen bond (red dashed lines) forming residues are shown in grey.
Docking study revealed that amentoflavone sufficiently gain some access through the hydrophobic channels of 15-lipoxygenase having binding energy of -10.8 kcal/mol (Table 4). The energy value suggests that the biflavonoid has very high affinity to the protein active site. Interestingly, this result is consistent with the potency (IC50 0.04 μM) with 85.3% inhibition earlier observed for the compound in wet experiment [43]. The biflavonoid is thus a strong competitive inhibitor of human 15-lipoxygenase and could be a desirable agent in the treatment of diseases associated with the protein functions. The inhibitory potential is higher than that of the reference ligand (pyrimido[4,5-b][1,4] benzothiazine derivative) as given in Table 5 having binding energy -7.5 (kcal/mol) with IC50 18 μM [65]. By binding at substrate binding pocket, the biflavonoid can disrupt substrate access, binding and conversion and hence, enzymatic catalysis. Three glutamine residues, Gln413, Gln363 and Gln609, participated in forming hydrogen bond with the compound in the active site. An essential π-π stacking with residue Phe177 and, hydrophobic interactions with Asn180, Leu607, Phe177 and Ala410 were also observed (Figure 4d). Another important observation is the interference of amentoflavone with the Fe2+ ion at the active site. The nonheme iron has been reported to be well conserved in lipoxygenases which possibly suggests its significance in enzyme catalysis for the protein [62,66]. Some inhibitors of this protein are known to chelate the iron molecules within the putative substrate binding site while others can convert Fe (III) to Fe (II). These mechanisms of lipoxygenase inhibition have been robustly investigated in human neutrophils and in isolated lipoxygenase systems [67-69].
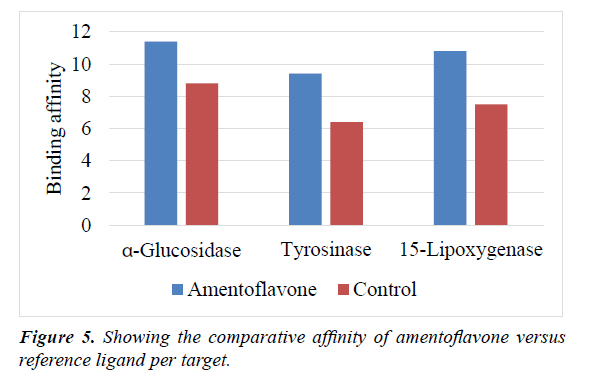
The inhibition associated with the formation of chelate-iron complexes in human lipoxygenase is known to be strong and can only be reversed by adding back iron to the experimental model. Since this discovery, the Fe (III)-containing active site of lipoxygenases has been targeted by synthesizing novel iron chelators [70-72]. By establishing a single hydrogen bond with the Fe2+ ion, perhaps the amentoflavone can chelate, initiate Fe3+ to Fe2+ reduction or at least interfere with the availability of the iron for protein use during enzymatic function [72]. In this study, it is clear that amentoflavone showed higher affinity and accordingly, higher potency against the selected targets compared to all the reference ligands employed (Figure 5). Hence, the chemical structure of this biflavonoid may be a critical starting candidate for the development of potent pharmaceutical agents capable of improving human health. Furthermore, synthetic derivitization of amentoflavone might yield valuable novel optimized inhibitors for treatment of inflammation, diabetes, cancer and melanin-related diseases.
Conclusion
In conclusion, this study reports the interaction profile and inhibitory potential of amentoflavone on the biological functions of few of its molecular targets, including α-glucosidase, tyrosinase and 15-lipoxygenase. These proteins have been implicated in various human diseases. The acclaimed beneficial effects of amentoflavone in the treatment of some of the diseases necessitated exhaustive evaluation of its precise interaction pattern with the targets. The obtained results in the current research suggest that the biflavonoid could directly interact with the proteins at the active site via competitive inhibition type. The bulky structure of the biflavonid permits it to efficiently occupy the catalytic site and other regions (the solvent exposed areas) of the enzyme, blocking substrate access and binding. In addition, interference with essential metal ions at the active site of tyrosinase and lipoxygenase by amentoflavone either via chelation or coordinate-disruption could result in inhibition of the catalytic cycle of the enzymes. The apigenin dimer is a potent competitive inhibitor of human α-glucosidase and 15-lipoxygenase, however a moderate inhibitor of tyrosinase protein. Its affinity and potency is relatively higher than that of the control ligands. The binding pattern generated for the biflavonoid against the enzymes in this study provides explanation and insights into its potency (IC50) against the targets. Amentoflavone is validated as a useful candidate and its optimization might yield efficacious agents to be applied in human diseases where α-glucosidase, tyrosinase and 15-lipoxygenase have been validated as therapeutic targets.
Acknowledgement
The author wishes to acknowledge technical support and encouragement received from all researchers at the Centre for Bio-computing and Drug Development (CBDD), Adekunle Ajasin University, Akungba-Akoko, Ondo state, Nigeria.
References
- Lee J, Jung K, Woo E, et al. Docking study of biflavonoids, allosteric inhibitors of protein tyrosine phosphatase 1b. Bull. Korean Chem. Soc. 2008; 29(8):1479-1484.
- Yu S, Yan H, Zhang L, et al. A review on the phytochemistry, pharmacology, and pharmacokinetics of amentoflavone, a naturally-occurring biflavonoid. Molecules. 2017; 22:299.
- Thapa A, Woo ER, Chi EY.et al. Biflavonoids are superior to monoflavonoids in inhibiting amyloid-β toxicity and fibrillogenesis via accumulation of nontoxic oligomer-like structures. Biochemistry. 2011; 50:2445-2455.
- Chen J, Chang HW, Kim HP, et al. Synthesis of phospholipase A2 inhibitory biflavonoids. Bioorgoranic Med Chem. Lett. 2006; 16:2373-2375.
- Tulin A. The significance of flavonoids as a potential anti-tuberculosis compounds. RRJPTS. 2015; 3:1-17.
- Vanessa SG, Jaqueline PJ, Wagner AJ. et al. Morelloflavone and its semisynthetic derivatives as potential novel inhibitors of cysteine and serine proteases. J. Med. Plant Res. 2015; 9:426-434.
- Okigawa M, Hwa CW, Kawano N, et al. Biflavones in Selaginella species. Phytochemistry. 1971; 10:3286-3287.
- Bais S, Abrol N. Review on chemistry and pharmacological potential of amentoflavone. Curr. Res. Neurosci. 2016; 6(1):16-22.
- Couleri P, Nour M, Maciuk A. et al. Structure-activity relationship study of biflavonoids on the Dengue virus polymerase DENV-NS5 RdRp. Planta Med. 2013; 79:1313-1318.
- Ndongo JT, Issa ME, Messi AN, et al. Cytotoxic flavonoids and other constituents from the stem bark of Ochna schweinfurthiana. Nat. Prod. Res. 2015; 29:1684-1687.
- Yue SM, Kang WY. Lowering blood lipid and hepatoprotective activity of amentoflavone from Selaginella tamariscina in vivo. J. Med. Plants Res. 2011; 5:3007-3014.
- Abdallah HM, Almowallad FM, Esmat A, et al. Antiinflammatory activity of flavonoids from Chrozophora tinctoria. Phytochem Lett. 2015; 13:74-80.
- Lee JS, Lee MS, Oh WK, et al. Fatty acid synthase inhibition by amentoflavone induces apoptosis and antiproliferation in human breast cancer cells. Biol. Pharm. Bull. 2009; 32:1427-1432.
- Zheng XK, Su CF, Zhang L, et al. Anti-diabetic activity of amentoflavone in Selaginella tamariscina in diabetic mice. Chin. J. Exp. Tradit. Med. Formuae. 2013; 19:198-202.
- Zhang Z, Sun T, Niu JG, et al. Amentoflavone protects hippocampal neurons: Anti-inflammatory, antioxidative, and antiapoptotic effects. Neural Regen. Res. 2015; 10:1125-1133.
- Bais S, Gill NS, Rana N. Effect of Juniperus communis extract on reserpine induced catalepsy. Inventi Impact: Ethnopharmacol. 2014; 4:117-120.
- Songca SP, Sebothoma C, Samuel BB, et al. A biflavonoid and a carotenoid from Rhus leptodictya: Isolation, characterization and antibacterial properties. Afr. J. Biochem. Res. 2012; 6:172-178.
- Sakthivel KM, Guruvayoorappan C. Amentoflavone inhibits iNOS, COX-2 expression and modulates cytokine profile, NF- κB signal transduction pathways in rats with ulcerative colitis. Int. Immunopharmacol. 2013; 17:907-916.
- Fischer C, Mazzone M, Jonckx B, et al. FLT1 and its ligands VEGFB and PlGF: Drug targets for anti-angiogenic therapy? Nat. Rev. Cancer. 2008; 8:942-956.
- Ahn JS. Protein tyrosine phosphatase 1B inhibitory activity of amentoflavone and its cellular effect on tyrosine phosphorylation of insulin receptors. Biol. Pharm. Bull. 2007; 30:379-381.
- An J, Li Z, Dong Y, et al. Amentoflavone protects against psoriasislike skin lesion through suppression of NF-κB-mediated inflammation and keratinocyte proliferation. Mol. Cell Biochem. 2016; 413:87-95.
- Zhang J, Liu Z, Cao W, et al. Amentoflavone inhibits angiogenesis of endothelial cells and stimulates apoptosis in hypertrophic scar fibroblasts. Burns 2014; 40:922-929.
- Xu P, Jiang EJ, Wen SY, et al. Amentoflavone acts as a radioprotector for irradiated v79 cells by regulating reactive oxygen species (ROS), cell cycle and mitochondrial mass. Asian Pac. J. Cancer Prev. 2014; 15:7521-7526.
- Chaabi M, Antheaume C, Weniger B, et al. Biflavones of Decussocarpus rospigliosii as phosphodiesterases inhibitors. Planta Med. 2007; 73:1284-1286.
- Dell'Agli M, Galli GV, Bosisio E. Inhibition of cGMP-phosphodiesterase-5 by biflavones of Ginkgo biloba. Planta Medica. 2006; 72:468-470.
- Kimura Y, Ito H, Ohnishi R, et al. Inhibitory effects of polyphenols on human cytochrome P450 3A4 and 2C9 activity. Food Chem. Toxicol. 2010; 48:429-435.
- Wilsky S, Sobotta K, Wiesener N, et al. Inhibition of fatty acid synthase by amentoflavone reduces coxsackie virus B3 replication. Arch. Virol. 2012; 157:259-269
- Banerjee T, Valacchi G, Ziboh VA, et al . Inhibition of TNF alpha-induced cyclooxygenase-2 expression by amentoflavone through suppression of NFkappa B activation in A549 cells. Mol. Cell Biochem. 2012; 238:105-110.
- Hanrahan JR, Chebib M, Davucheron NLM, et al. Semisynthetic preparation of amentoflavone: A negative modulator at GABAA receptors. Bioorg. Med. Chem. Lett. 2003; 13:2281-2284.
- Zheng XK, Ning TL, Wang XL, et al. Effects of total flavonoids and amentoflavone isolated from Selaginella tamariscina on human umbilical vein endothelial cells proliferation and VEGF expression. Chin. Pharm. J. 2011; 13:998-1002.
- Pan X, Tan N, Zeng G, et al. Amentoflavone and its derivatives as novel natural inhibitors of human Cathepsin B. Bioorg. Med. Chem., 2005; 13:5819-5825.
- Laishram S, Sheikh Y, Moirangthem DS, et al. Anti-diabetic molecules from Cycas pectinata Griff. traditionally used by the Maiba-Maibi. Phytomed. 2015; 22:23-26.
- Ogunwa TH, Ayenitaju FC. An insight into the precise molecular interaction and inhibitory potential of amentoflavone and its substituted derivatives on human α-amylase. Arch. Curr. Res. Int., 2017; 10(1):1-14.
- Ogunwa TH, Ayenitaju FC. Molecular binding signatures of morelloflavone and its naturally occurring derivatives on HMG-COA reductase. Int. J. Biol. Sci. Appl., 2017b; 4(5):74-81.
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010; 31:455-461.
- Seelinger D, de Groot BL. Ligand docking and binding site analysis with PYMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010; 24:417-422.
- Stierand K, Maaß P, Rarey M. Molecular complexes at a glance: Automated generation of two-dimensional complex diagrams. Bioinformatics. 2006; 22(14):1710-1716.
- Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem. Inf. Model. 2011; 51(10):2778-86.
- Ali F, Rahul, Naz F, et al. Health functionality of apigenin: A review. Int. J. Food Proper. 2017; 20(6):1197-1238.
- Kim HP, Son KH, Chang HW, et al. Anti-inflammatory plant flavonoids and cellular action mechanisms. J. Pharmacol. Sci. 2004; 96: 229-245.
- Katavic PL, Lamb K, Navarro H, et al. Flavonoids as opioid receptor ligands: Identification and preliminary structure–activity relationships. J. Nat. Prod. 2007; 70(8):1278-1282.
- Wasowski C, Marder M. Flavonoids as GABAA receptor ligands: the whole story? J Exp Pharmacol. 2012; 4:9-24.
- Aminudina NI, Ahmada F, Taherb M, et al. α-glucosidase and 15-lipoxygenase inhibitory activities of phytochemicals from calophyllum symingtonianum. Nat. Prod. Comm. 2015; 10(9):1585-1587
- Loizzo MR, Tundis R, Menichini F. Natural and Synthetic Tyrosinase Inhibitors as Antibrowning Agents: An Update. Comp. Rev. Food Sci. Food Safety, 2012; 11:378-398.
- Bischoff H. The mechanism of alpha-glucosidase inhibition in the management of diabetes. Clin. Invest. Med. 1995; 18(4):303-311.
- Rossi, E. J., Sim, L., Kuntz, D. A., et al. Inhibition of recombinant human maltase glucoamylase by salacinol and derivatives. FEBS J. 2006; 273: 2673–2683.
- Kim JS, Kwon CS, Son KH. Inhibition of alpha-glucosidase and amylase by luteolin, a flavonoid. Biosc. Biotechnol. Biochem. 2000; 64:2458-2461.
- Luthra T, Agarwal R, Estari M, et al., A novel library of –arylketones as potential inhibitors of α-glucosidase: Their design, synthesis, in vitro and in vivo studies. Scientific reports 2017; 7:13246
- Qin H, Shang Z, Jantan I, et al. Molecular docking studies and biological evaluation of chalcone based pyrazolines as tyrosinase inhibitors and potential anticancer agents. RSC Adv 2015; 5: 46330-46338
- Quispe YN, Hwang SH, Wang Z, et al. Screening of peruvian medicinal plants for tyrosinase inhibitory properties: identification of tyrosinase inhibitors in Hypericum laricifolium Juss. Molecules. 2017; 22: 402.
- Arianayagam S, Ryan TJ. Human pigmentation: A side effect adapted from a primitive organism's survival. Part 2: The melanocyte as mentor of the keratinocyte. Indian Dermatol. Online J. 2014; 5(3):328-333.
- Olivares C, Solano F. New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins. Pigment Cell Melanoma Res. 2009; 22(6):750-760.
- Ashraf Z, Rafiq M, Seo S. Kinetic and in silico studies of novel hydroxy-based thymol analogues as inhibitors of mushroom tyrosinase. Eur. J. Med. Chem. 2015; 98:203-211.
- Ismaya WT, Rozeboom HJ, Weijn A,et al. Crystal structure of Agaricus Bisporus mushroom tyrosinase: identity of the tetramer subunits and interaction with tropolone. Biochemistry. 2011; 50(24):5477-5486.
- Kahn V, Andrawis A. Inhibition of mushroom tyrosinase by tropolone. Phytochem. 1985; 24(5): 905-908.
- Asadzadeh A, Fassihi A, Yaghmaei P, et al. Docking studies of some novel kojic acid derivatives as possible tyrosinase inhibitors. Biomed. Pharmacol. J. 2015; 8(2): 535-545.
- Asadzadeh A, Sirous H, Pourfarzam M, et al. In vitro and in silico studies of the inhibitory effects of some novel kojic acid derivatives on tyrosinase enzyme. Iran J. Basic Med. Sci. 2016; 19:132-144.
- Rescigno A, Sollai F, Pisu B, et al. Tyrosinase inhibition: general and applied aspects. J Enz. Inhib. Med. Chem. 2002; 17(4): 207-218.
- Roy SK, Qasim MA, Kamil M, et al. Biflavones from the genus Podocarpus. Phytochem. 1987; 26:1985-1987.
- Cheng KT, Hsu FL, Chen SH, et al. New constituent from Podocarpus macrophyllus var. macrophyllus shows anti-tyrosinase effect and regulates tyrosinase-related proteins and mRNA in human epidermal melanocytes. Chem. Pharm. Bull. 2007; 55(5):757-761.
- Wecksler AT, Kenyon V, Garcia NK, et al. Kinetic and structural investigations of the allosteric site in human epithelial 15-lipoxygenase-2. Biochemistry. 2009; 48: 8721-8730.
- Mogul R, Theodore R. Inhibition studies of soybean and human 15-lipoxygenases with long-chain alkenyl sulfate substrates. Biochemistry. 2001; 40:4391-4397.
- Cornicelli J, Trivedi BK. 15-Lipoxygenase and its inhibition: a novel therapeutic target for vascular disease. Curr. Pharm. Des. 2010; 5(1):11-20.
- Wisastra R, Dekker FJ. Inflammation, cancer and oxidative lipoxygenase activity are intimately linked. Cancers. 2014; 6: 1500-1521.
- Neau DB, Bender G, Boeglin WE, et al. Crystal structure of a lipoxygenase in complex with substrate: the arachidonic acid-binding site of 8R-lipoxygenase. J. Biol. Chem. 2014; 289(46):31905-31913.
- Bakavoli M, Nikpour M, Rahimizadeh M, Saberi MR, Sadeghian H. Design and synthesis of pyrimido[4,5-b][1,4]benzothiazine derivatives, as potent 15-lipoxygenase inhibitors. Bioorg Med Chem. 2007;15(5):2120-2126.
- Rossaint J, Nadler JL, Ley K, et al. Eliminating or blocking 12/15-lipoxygenase reduces neutrophil recruitment in mouse models of acute lung injury. Crit Care. 2012; 16(5) R166.
- Eleftheriadis N, Dekker FJ. The role of human 15-lipoxygenase-1 in Asthma. SM J. Pulm. Med. 2016; 2(1):1015.
- Kwon HJ, Kim SN, Kim YA, et al. The contribution of arachidonate 15-lipoxygenase in tissue macrophages to adipose tissue remodeling. Cell Death and Disease. 2016; 7: e2285;
- Kobe MJ, Neau DB, Mitchell CE, et al. The structure of human 15-lipoxygenase-2 with a substrate mimic. The J. Biol. Chem. 2014; 289(12):8562-8569,
- Skaterna TD, Kopich VM, Kharitonenko GI, et al. Lipoxygenase regulation in vivo and in vitro by lipid compounds. Biopolymers and Cell. 2015; 31(3):161-173.
- Sadeghian H, Jabbari A. 15-Lipoxygenase inhibitors: a patent review, Expert Opinion on Therapeutic Patents, 2015; 26(1): 65-88.