Research Article - Biomedical Research (2017) Volume 28, Issue 18
Inhibitory effect of microRNA-605-3p on biological functions of melanoma A375 cells via wnt11
Shoujie Wang1 and Yandong Li2*
1Department of Plastic Surgery, the First Affiliated Hospital, Zhejiang University, Hangzhou, PR China
2Department of General Surgery, the First Affiliated Hospital, Zhejiang University, Hangzhou, PR China
- *Corresponding Author:
- Yandong Li
Department of General Surgery
The First Affiliated Hospital, Zhejiang University, PR China
Accepted on August 28, 2017
Abstract
Aims: The aim of the present study is to examine microRNA (miRNA or miR)-605-3p expression in melanoma at tissue and cellular levels, and to understand its biological roles.
Methods: A total of 45 melanoma patients (27 males and 18 females) were enrolled in this research. To measure the expression of miR-605-3p, quantitative real-time polymerase chain reaction was employed. Melanoma A375 cells were transfected with miR-605-3p mimics or inhibitor. CCK-8 assay was used to investigate the proliferation of a375 cells. To study migration and invasion abilities of A375 cells, Transwell assay was carried out. To determine the expression of wnt11 protein, we used Western blotting. Rescue assay was utilized to study the biological roles in A375 cells. To test whether wnt11 is the direct target of miR-605-3p, dual luciferase reporter assay was performed.
Results: Expression of miR-605-3p in melanoma tissues was decreased, and negatively correlated with lymph node metastasis and clinical staging. In vitro, expression of miR-605-3p inhibited the proliferation of melanoma A375 cells. Moreover, expression of miR-605-3p suppressed migration and invasion abilities of melanoma A375 cells. Interestingly, miR-605-3p regulated the expression of wnt11 protein in A375 cells. Of note, wnt11 promoted the proliferation, migration and invasion abilities of melanoma A375 cells. Dual luciferase reporter assay showed that miR-605-3p was able to bind with the 3’-untranslated region seed region of wnt11 mRNA to regulate its expression.
Conclusion: The present study shows that miR-605-3p expression is decreased in melanoma tissues. In addition, miR-605-3p is negatively correlated with lymph node metastasis and clinical staging. As a tumor-suppressor gene, miR-605-3p inhibits the proliferation, migration and invasion of melanoma A375 cells by suppressing the expression of wnt11.
Keywords
microRNA-605-3p, Melanoma, wnt11.
Introduction
Melanoma is a kind of clinically common tumor with high degree of malignancy and poor prognosis, and it is usually named as Malignant Melanoma (MM) [1,2]. MM is common in the skin, accounting for 10% of skin cancers, and its mortality rate reaches up to 80% [3]. In recent years, MM has shown increasing incidence and familial aggregation [4]. It is difficult to carry out early diagnosis and treatment for MM, because its early clinical symptoms are usually concealed. Most MM patients already have local lymph node or distant metastases at the first diagnosis, and the disease rapidly progresses after diagnosis, finally leading to death [5]. It is generally accepted that melanoma is derived from melanocytes in the basal layer of epidermis, and has a wide range of tight junctions [6]. However, it is still unclear how tumor cells break this type of junctions, dissociate from their primary sites, and initiate invasion and metastasis.
MicroRNA (miRNA or miR) is a type of non-coding small RNA with 18-22 nucleotides. As a kind of post-transcriptional regulator, miRNA molecules form protein silencing complex after binding to 3’-untranslated region (UTR) of mRNA, inhibit the translation of mRNA, and regulate protein expression [7,8]. miRNA is found to play important roles in the regulation of cell proliferation, migration, apoptosis, autophagy and differentiation [9,10]. It has been found that miRNA plays the roles of oncogenes or tumor-suppressor genes in tumorigenesis [11]. Different miRNA molecules play distinct biological roles in tumor, and these molecules have tissue specificity [12]. miRNA plays important roles in the proliferation, metastasis and drug resistance of melanoma. For example, miR-204-5p inhibits occurrence and development of melanoma by regulating Matrix Metalloproteinase (MMP)-9 and B-Cell Lymphoma-2 (BCL2) levels [13]. Sun et al. report that proliferation and metastasis of melanoma is decreased by miR-216b through targeting FOXM1 gene [14]. These studies indicate that miRNA molecules may be the therapeutic targets for melanoma.
In recent years, the wide application of microarray technology, especially gene chip, has provided a large amount of highthroughput data to studying changes of miRNA expression in tumors and to screen miRNA molecules that are related to tumor diagnosis and therapy [15]. Geo Datasets, owned by National Center for Biotechnology Information of National Institutes of health, is the largest and the most comprehensive public gene chip database [16]. Using this database, we have analysed miRNA expression changes in melanoma (GSE No.: GSE8401), and discovered that miR-605-3p expression is significantly reduced in metastatic melanoma tissues, suggesting that miR-605-3p plays important roles in the invasion and migration of melanoma cells. Here, we examine miR-605-3p expression in melanoma at tissue and cellular levels, and investigate its biological roles in melanoma.
Materials and Methods
Patients
A total of 45 melanoma patients (27 males and 18 females) who received diagnosis and treatment at The First Affiliated Hospital of Zhejiang University between July 2014 and July 2016 were enrolled in the research. Their age range was 35-71 y, and their average age was 51.5 y. Patients with lymph node metastasis (27 cases) were included into N1 subgroup, while those without lymph node metastasis (18 cases) were included into N0 subgroup. According to Clark classification for infiltration degree, 11 patients were classified into grade I (epidermis and adnexal epithelia), 17 patients were classified into grades II and III (papillary layer of dermis), and 8 patients were classified into grade IV (reticular layer of dermis), and 9 patients were classified into grade V (subcutaneous layer). In addition, 26 skin grafts left over from plastic surgery at our hospital were used as controls. All procedures were approved by the Ethics Committee of Zhejiang University. Written informed consents were obtained from all patients or their families.
Cells
Melanoma A375 cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% Fetal Bovine Serum (FBS), 100 IU/ml penicillin and 100 IU/ml streptomycin under 37°C, 5% CO2, and 70% humidity. A375 cells were passaged every four days, and logphase cells were collected for experiments.
On the day before transfection, A375 cells (2 × 105) in logphase growth were inoculated into 24-well plates, and cultured in serum-free RPMI-1640 medium until reaching 70% confluency. In the first vial, 1.5 μL miR-605-3p inhibitor (20 pmol/μL; inhibitor group) or mimics (20 pmol/μL; mimics group) (Hanbio Biotechnology Co., Ltd., Shanghai, China) was mixed gently with 50 μl Opti Mem medium (Thermo Fisher Scientific, Waltham, MA, USA). In the second vial, 1 μL Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was mixed gently with 50 μl Opti Mem medium. After five minutes, the two vials were combined for another waiting at room temperature for twenty minutes. Then, the mixtures were added onto cells in respective groups. Six hours later, the medium was replaced with RPMI-1640 medium containing 10% FBS. After being incubated for 48 h, these cells were collected for further assays.
For rescue experiments, A375 cells (2 × 105) in miR-605-3p mimics or inhibitor group were seeded into 24-well plates containing antibiotics-free RPMI-1640 medium supplemented with 10% FBS. When reaching 60% confluency, A375 cells in miR-605-3p mimics or inhibitor groups were infected by Lvpuro- sh-wnt11 or Lv-puro-wnt11 lentiviral vectors (MOI=20; Hanbio Biotechnology Co., Ltd., Shanghai, China). After being cultured at 37°C and under 5% CO2 for 12 h, the medium was refreshed to newly made RPMI-1640 medium containing 1 μg/ml puro before cultivation for 72 h.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Melanoma and control tissues (100 mg) were frozen and made into powder in liquid nitrogen, and then mixed with Trizol reagent (1 ml) for lysis. After that, phenol chloroform method was employed for the extraction of total RNA. To test the purity of extracted RNA, ultraviolet spectrophotometry was used (Nanodrop ND2000, Thermo Scientific, Waltham, MA, USA). Then, 1 μg RNA was reverse-transcribed into cDNA that was stored at -20°C. Reverse transcription of was carried out using miScript II RT Kit (Qiagen, Hilden, Germany).
The expression of miRNA was determined by miScript SYBR® Green PCR Kit (Qiagen, Hilden, Germany), using U6 as internal reference. Reaction system (20 μl) contained qRTPCR- Mix (10 μl), upstream primer (0.5 μl; miR-106b, 5’- AGAAGGCACUAUGAGAUUUAGA-3’), downstream universal primer (0.5 μl; provided by the kit), cDNA (2 μl) and ddH2O (7 μl). After initial denaturation for 10 min at 95°C, the samples were subjected to 40 cycles of incubation at 95°C (1 min) and 60°C (30 s) (iQ5; Bio-Rad, Hercules, CA, USA). To calculate relative expression of miR-605-3p, 2-ΔΔCt method was performed against the internal reference. Every sample was examined in three replicates.
CCK-8 assay
The sample cells were inoculated in 96-well plates at a density of 2,000/well. At 0 h, 24 h, 48 h and 72 h, 20 μl CCK-8 (5 g/L) was added onto the cells. On the last day, 150 μl CCK-8 reaction liquid was added onto the cells, which were subsequently incubated at 37°C for 2 h. Absorbance (490 nm) of each well was determined, and cell proliferation curves were plotted. Each group was tested in 3 replicate wells and the values were averaged.
Transwell assay
Matrigel was defrosted at 4°C overnight and diluted with serum-free RPMI-1640 medium at a dilution ratio of 1:2. The mixture (50 μl) was evenly smeared into upper chamber (Merck Millipore, Billerica, MA, USA). After being incubated at 37°C for 1 h and solidification, cells (1 × 105) in each group were inoculated into upper chamber that already contained 200 μl serum-free RPMI-1640 medium. Subsequently, 500 μl RPMI-1640 medium containing 10% FBS was added into lower chamber. Twenty-four hours later, cells in upper chamber were wiped off and the upper chamber was fixed with 4% formaldehyde for 10 min. Then, Giemsa method was used to stain the membrane, which was later observed under a microscope in 5 random fields (magnification, 200X). The number of Transwell cells was counted in order to evaluate cell invasion ability and migration ability. All these steps were performed on ice with precooled pipetting tips (4°C).
Western blotting
Cells in each group were trypsinized and collected. Then, cold Radio-Immunoprecipitation Assay (RIPA) lysis buffer (600 μl; Beyotime Institute of Biotechnology, Shanghai, China) was mixed with the samples. Then, the mixture was lysed for 30 min on ice, and then centrifuged at 12,000 rpm and 4°C for 10 min. Bicinchoninic Acid (BCA) protein concentration determination kit (RTP7102, Real-Times Biotechnology Co., Ltd., Beijing, China) was used to determine protein concentration in the supernatant. After mixing protein samples (6 μl) with 5X sodium dodecyl sulfate loading buffer, the mixture was denatured by boiling in water bath for 10 min. Afterwards, 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (100 V) was performed using the samples. Then, the proteins were electro-transferred to Polyvinylidene Difluoride (PVDF) membranes on ice (250 mA, 1 h) before being blocked with skimmed milk (50 g/L) at room temperature for 1 h. Afterwards, rabbit anti-human wnt11 polyclonal primary antibody (1:1,000; Santa Cruz, Dallas, TX, USA) and rabbit anti-human GAPDH primary antibody (1:4,000; Santa Cruz, Dallas, TX, USA) were added onto the membranes before incubation at 4°C overnight. Then, the membrane was extensively washed with Phosphate-Buffered Saline with Tween 20 (PBST), and incubated with goat antirabbit horseradish peroxidase-conjugated secondary antibody (1:2,000; Santa Cruz, Dallas, TX, USA) at room temperature for 1 h. Subsequently, the membrane was washed with PBST for 5 times of 5 min before the membrane was developed with enhanced chemiluminescence detection kit (Sigma-Aldrich, St. Louis, MO, USA) for imaging. We used Image lab v3.0 software (Bio-Rad, Hercules, CA, USA) to acquire and analyse imaging data. The relative expression of wnt11 protein was expressed with the ratio of wnt11/GAPDH.
Dual luciferase reporter assay
Wild-Type (WT) and mutant seeding regions of miR-605-3p in 3’-UTR of wnt11 gene were chemically synthesized before adding Spe-1 and HindIII restriction sites, and then cloned into pMIR-REPORT luciferase reporter plasmids (0.5 μg) with WT or mutant 3’-UTR DNA sequences, which were transfected together with miR-605-3p mimics into HEK293T cells. Following incubation for 24 h, cells were processed using dual luciferase reporter assay kit according to the manufacturer’s manual (Promega, Fitchburg, WI, USA), and fluorescence intensity was determined by GloMax 20/20 luminometer (Promega, Fitchburg, WI, USA). Fluorescence values of each group of were measured using renilla fluorescence activity as internal reference.
Statistical analysis
All results were analysed using SPSS 17.0 statistical software (IBM, Armonk, NY, USA), and all data were shown as means ± SD. Comparison between groups was performed using group t-test. Differences with P<0.05 were considered statistically significant.
Results
miR-605-3p expression in melanoma tissues is decreased, and negatively correlated with lymphatic metastasis and clinical staging
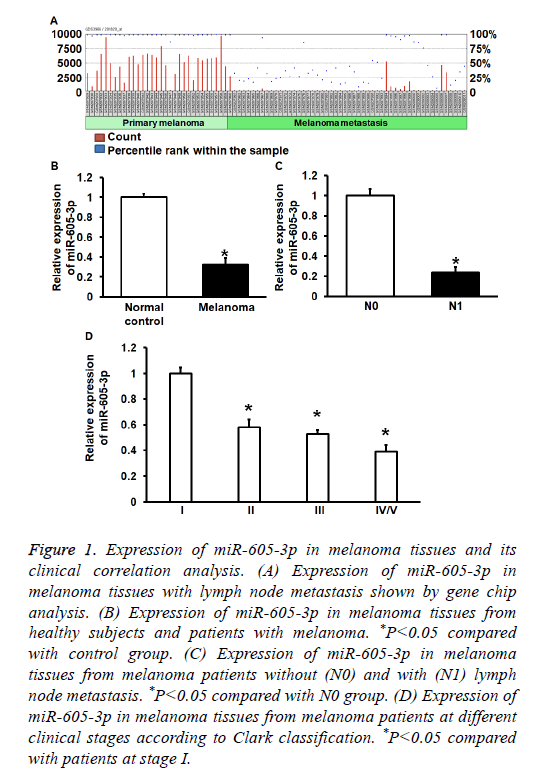
To determinemiR-605-3p expression in melanoma tissues and A375 cells, GEO Datasets and qRT-PCR were used. GEO Datasets (GSE8401) analysis showed that miR-650-3p expression in melanoma with lymphatic metastasis was significantly reduced compared with that in melanoma without lymphatic metastasis (P<0.01) (Figure 1A). qRT-PCR data showed that miR-605-3p expression in melanoma tissues was significantly decreased than that in controls (P<0.01) (Figure 1B). In addition, miR-605-3p expression in melanoma tissues from patients with lymphatic metastasis was significantly reduced than that in tissues from patients without lymph node metastasis (P<0.01) (Figure 1C). Moreover, miR-605-3p expression in stage IV/V melanoma tissues was significantly decreased than that in stages I-III (P<0.05) (Figure 1D). The data suggest that miR-605-3p expression in melanoma tissues is decreased, and negatively correlated with lymph node metastasis and clinical staging.
Figure 1: Expression of miR-605-3p in melanoma tissues and its clinical correlation analysis. (A) Expression of miR-605-3p in melanoma tissues with lymph node metastasis shown by gene chip analysis. (B) Expression of miR-605-3p in melanoma tissues from healthy subjects and patients with melanoma. *P<0.05 compared with control group. (C) Expression of miR-605-3p in melanoma tissues from melanoma patients without (N0) and with (N1) lymph node metastasis. *P<0.05 compared with N0 group. (D) Expression of miR-605-3p in melanoma tissues from melanoma patients at different clinical stages according to Clark classification. *P<0.05 compared with patients at stage I.
Expression of miR-605-3p inhibits the proliferation of melanoma A375 cells
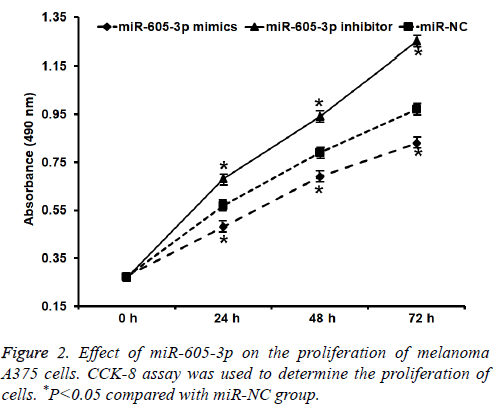
To test the proliferation of A375 cells, CCK-8 assay was carried out. These data showed that absorbance of A375 cells that were transfected with miR-605-3p mimics was significantly reduced than that in miR-NC group at all-time points (P<0.05), while the absorbance of A375 cells in miR-605-3p inhibitor group was significantly increased than that in miR-NC group at all-time points (P<0.05) (Figure 2). The result indicates that miR-605-3p expression inhibits melanoma A375 cell proliferation.
Expression of miR-605-3p inhibits migration and invasion abilities of melanoma A375 cells
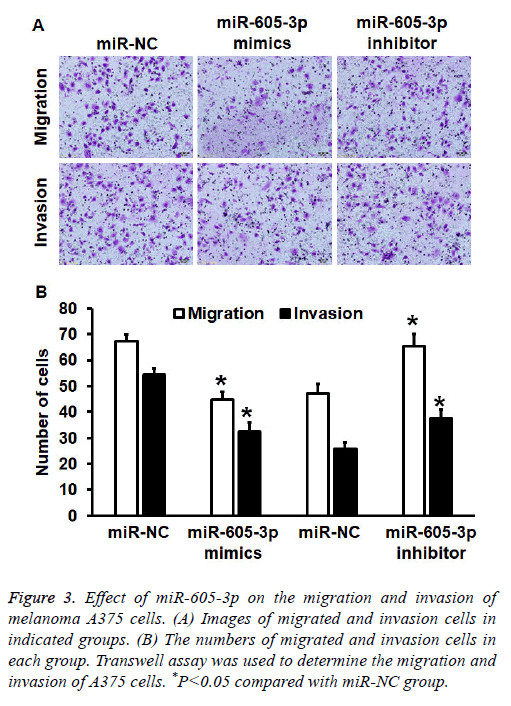
To investigate migration and invasion abilities of A375 cells, Transwell assay was employed. The data showed that the number of cells in miR-605-3p mimics group that crossed chamber membrane in migration assay was reduced than that in miR-NC group (P<0.05). By contrast, cell count in miR-605-3p inhibitor group that crossed chamber membrane in migration assay was higher than that in miR-NC group (P<0.05). Similarly, cell count in miR-605-3p mimics group that crossed chamber membrane in invasion assay was reduced than that in miR-NC group (P<0.05). In addition, the number of cells in miR-605-3p inhibitor group that crossed chamber membrane in invasion assay was higher than that in miR-NC group (P<0.05) (Figures 3A and 3B). The results suggest that miR-605-3p expression inhibits migration and invasion abilities of melanoma A375 cells.
Figure 3: Effect of miR-605-3p on the migration and invasion of melanoma A375 cells. (A) Images of migrated and invasion cells in indicated groups. (B) The numbers of migrated and invasion cells in each group. Transwell assay was used to determine the migration and invasion of A375 cells. *P<0.05 compared with miR-NC group.
Expression of wnt11 protein is regulated by miR-605-3p in A375 cells
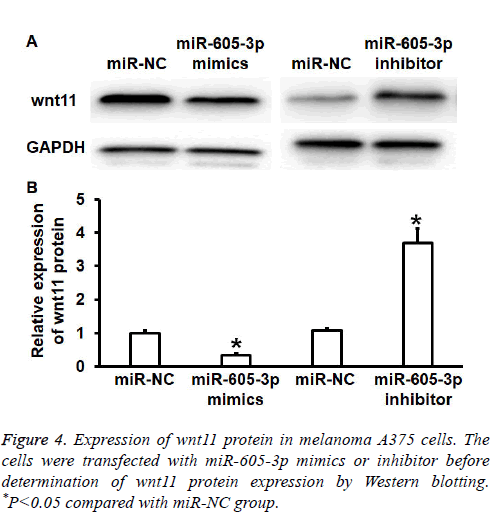
To test wnt11 protein expression in A375 cells, Western blotting was performed. Quantification of Western blots showed that wnt11 protein expression in miR-605-3p mimics group was significantly reduced than that in miR-NC group (P<0.05), while wnt11 protein expression in miR-605-3p inhibitor group was significantly increased than that in miRNC group (P<0.05) (Figures 4A and 4B). The result suggests that miR-605-3p regulates the expression of wnt11 protein in A375 cells.
Wnt11 promotes proliferation, migration and invasion abilities of melanoma A375 cells
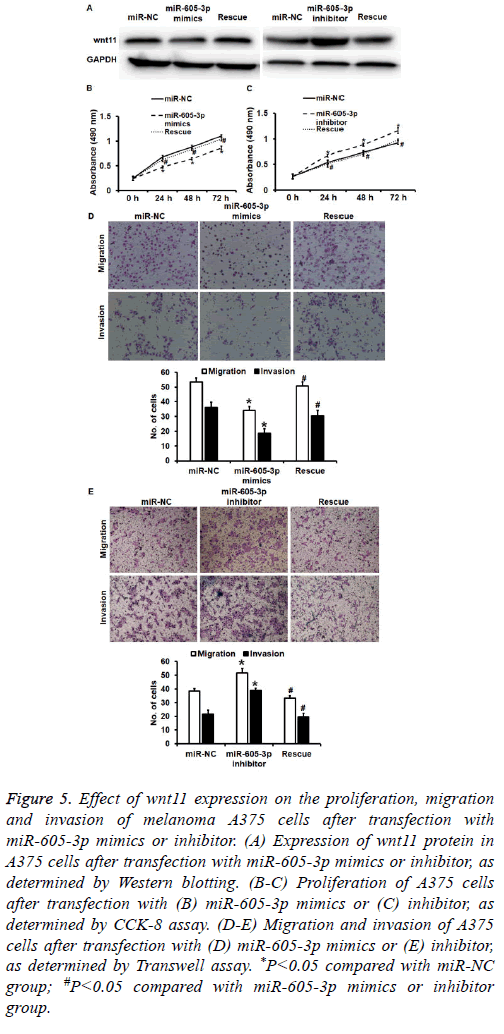
To test how the regulation of wnt11 by miR-605-5p affects biological functions of A375 cells, we silenced or overexpressed wnt11 protein using Lv-puro-sh-wnt11 or Lvpuro- wnt11 lentiviral vectors. Western blotting showed that expression of wnt11 in miR-605-5p mimics or inhibitor groups was rescued (Figure 5A). CCK-8 assay showed that downregulation of wnt11 induced by overexpression of miR-605-5p decreased the proliferation of A375 cells (P<0.05) (Figure 5B), while overexpression of wnt11 induced by inhibition of miR-605-5p promoted the proliferation of A375 cells (P<0.05) (Figure 5C). Transwell assay data showed that down-regulated wnt11 expression induced by overexpression of miR-605-5p reduced transwell cell count in both migration and invasion assays compared with that in miR-NC group (P<0.05) (Figure 5D), while overexpression of wnt11 induced by inhibition of miR-605-5p increased transwell cell count in both migration and invasion assays compared with that in miR-NC group (P<0.05) (Figure 5E). The results indicate that wnt11 promotes the proliferation, migration and invasion of melanoma A375 cells.
Figure 5: Effect of wnt11 expression on the proliferation, migration and invasion of melanoma A375 cells after transfection with miR-605-3p mimics or inhibitor. (A) Expression of wnt11 protein in A375 cells after transfection with miR-605-3p mimics or inhibitor, as determined by Western blotting. (B-C) Proliferation of A375 cells after transfection with (B) miR-605-3p mimics or (C) inhibitor, as determined by CCK-8 assay. (D-E) Migration and invasion of A375 cells after transfection with (D) miR-605-3p mimics or (E) inhibitor, as determined by Transwell assay. *P<0.05 compared with miR-NC group; #P<0.05 compared with miR-605-3p mimics or inhibitor group.
miR-605-3p can bind with the 3’-UTR seed region of wnt11 mRNA to regulate its expression
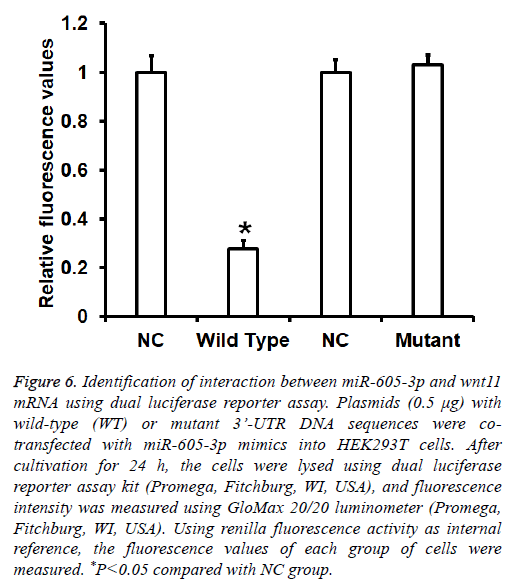
To identify the interaction between miR-605-3p and the 3’- UTR of wnt11 mRNA, dual luciferase reporter assay was performed. The fluorescence value of cells co-transfected with miR-605-3p mimics and pMIR-REPORT-WT luciferase reporter plasmids was significantly lower than that in negative control group (P<0.05). By contrast, the fluorescence value of cells co-transfected with miR-605-3p mimics and pMIRREPORT- mutant luciferase reporter plasmids was not significantly different from that in NC group (P>0.05) (Figure 6). The result indicates that miR-605-3p can bind with the 3’- UTR seeding region of wnt11 mRNA to regulate its expression.
Figure 6: Identification of interaction between miR-605-3p and wnt11 mRNA using dual luciferase reporter assay. Plasmids (0.5 μg) with wild-type (WT) or mutant 3’-UTR DNA sequences were cotransfected with miR-605-3p mimics into HEK293T cells. After cultivation for 24 h, the cells were lysed using dual luciferase reporter assay kit (Promega, Fitchburg, WI, USA), and fluorescence intensity was measured using GloMax 20/20 luminometer (Promega, Fitchburg, WI, USA). Using renilla fluorescence activity as internal reference, the fluorescence values of each group of cells were measured. *P<0.05 compared with NC group.
Discussion
Recurrence and metastasis are the main causes of poor prognosis in melanoma patients. Studies show that the migration of melanoma cells is regulated by multiple genes and factors, such as disorder of gene expression, aberrant methylation, gene mutation, tumor microenvironment changes [17,18]. Microarray data indicates that the expression of miRNA in melanoma cells is disordered, suggesting that miRNA molecules are important in the occurrence and development of melanoma [19]. Here, we find that miR-605-3p expression is significantly down-regulated in melanoma tissues and negatively correlated with the clinical staging and lymphatic metastasis of melanoma, suggesting that miR-605-3p participates in the occurrence and development of melanoma. In vitro experiments show that miR-605-3p inhibits proliferation, migration and invasion abilities of melanoma A375 cells by directly targeting wnt11. These results demonstrate that miR-605-3p has the function of tumorsuppressor genes, and may be a therapeutic target for tumors.`
Expression of various miRNA molecules is disordered in melanoma tissues, and these miRNA molecules widely participate in the proliferation, migration, invasion, apoptosis and drug resistance of tumor cells [20]. For example, Zhang et al. discover that miR-4262 promotes the proliferation, metastasis and invasion of melanoma by regulating the deactivation of EGFR and up-regulation of p21that are mediated by KLF6 [21]. Qiu et al. report that miR-186 enhances the proliferation of melanoma cells by inhibiting CYLD expression [22]. Sun et al. show that miR-216b suppresses proliferation and migration abilities of melanoma cells by targeting FOXM1 [14]. In the present study, we find that miR-605-3p expression is down-regulated in melanoma tissues, and negatively correlated with lymph node metastasis. Further analysis shows that miR-605-3p levels in stage IV/V melanoma tissues are significantly lower than those at stages IIII, suggesting that miR-605-3p expression is negatively correlated with the clinical staging of melanoma. At cellular level, we discover that silencing of miR-605-3p expression promotes proliferation, migration and invasion abilities of melanoma A375 cells, while miR-605-3p overexpression inhibits proliferation, migration and invasion abilities of A375 cells. The results indicate that miR-605-3p plays a role as an oncogene, and up-regulation of miR-605-3p promotes the proliferation, metastasis and invasion abilities of melanoma.
As the initial protein of the wnt signaling pathway, wnt11 is first discovered to control tissue morphogenesis [23]. Wnt11 is not only related to individual development, cell adhesion, migration, proliferation, differentiation and death, but also closely related to tumorigenesis. It can act as oncogene or tumor-suppressor gene [24]. Wnt11 activates its downstream receptors when mediating wnt signaling pathway, thereby affecting the biological functions of cells. It is found that wnt11 participates in the regulation of proliferation and metastasis of various tumors. For example, Liu et al. discover that the expression of wnt11 is positively correlated with lymph node metastasis and TNM stage in esophageal squamous cell carcinoma patients [25]. Wei et al. show that wnt11 promotes invasion of cervical cancer cells [26]. According to bioinformatics analysis, WNT11 gene is one of the potential target genes of miR-605-3p. Our results in rescue experiments show that WNT11 overexpression restores the proliferation, metastasis and invasion of A375 cells transfected with miR-605-3p mimics, while silencing of WNT11 expression decreases the proliferation, migration and invasion of A375 cells transfected with miR-605-3p inhibitor. The data indicate that miR-605-3p inhibits the occurrence and development of melanoma by down-regulating the expression of WNT11. Western blotting and dual luciferase reporter assay confirm that miR-605-3p is able to bind with the 3’-UTR region of WNT11 mRNA, and down-regulate WNT11 protein expression. These results demonstrate that WNT11 is a target gene of miR-605-3p.
In conclusion, miR-605-3p inhibits the proliferation, metastasis and invasion of melanoma by down-regulating WNT11 expression, and is a potential molecular therapeutic target and marker
Acknowledgements
This work was supported by The First Affiliated Hospital, Zhejiang University. We would also like to thank Dr. Weifang Zhu for providing technical instructions.
Disclosures
All authors declare no financial competing interests. All authors declare no non-financial competing interests.
References
- Chhabra G, Ndiaye MA, Garcia-Peterson LM, Ahmad N. Melanoma chemoprevention: current status and future prospects. Photochem Photobiol 2017.
- Carrera C, Gual A, Diaz A, Puig-Butille JA, Nogues S, Vilalta A, Conill C, Rull R, Vilana R, Arguis P, Vidal-Sicart S, Alos L, Palou J, Castel T, Malvehy J, Puig S. Prognostic role of the histological subtype of melanoma on the hands and feet in Caucasians. Melanoma Res 2017.
- Guo W, Wang H, Yang Y. Down-regulated miR-23a contributes to the metastasis of cutaneousmelanoma by promoting autophagy. Theranostics 2017; 7: 2231-2249.
- Gladfelter P, Darwish NHE, Mousa SA. Current status and future direction in the management of malignant melanoma. Melanoma Res 2017.
- Su Y, Hu J, Huang Z, Huang Y, Peng B, Xie N, Liu H. Paclitaxel-loaded star-shaped copolymer nanoparticles for enhanced malignant melanoma chemotherapy against multidrug resistance. Drug Des Devel Ther 2017; 11: 659-668.
- Tas F, Bilgin E, Erturk K, Duranyildiz D. Clinical significance of serum claudin-1 levels in melanoma patients. Melanoma Res 2016; 26: 377-381.
- Zhang R, Li R, Lin Y. Identification and characterization of microRNAs in the muscle of Schizothorax prenanti. Fish Physiol Biochem 2017.
- Kao SC, Cheng YY, Williams M. Tumor Suppressor microRNAs Contribute to the Regulation of PD-L1 Expression in malignant pleural mesothelioma. J Thorac Oncol 2017; 17; 30460-30464.
- Chou CK, Liu RT, Kang HY. MicroRNA-146b: A novel biomarker and therapeutic target for human papillary thyroid cancer. Int J Mol Sci 2017; 18.
- Bai L, Wang H, Wang AH, Zhang LY, Bai J. MicroRNA-532 and microRNA-3064 inhibit cell proliferation and invasion by acting as direct regulators of human telomerase reverse transcriptase in ovarian cancer. PloS One 2017; 12: 0173912.
- Wang H, Tang C, Na M, Ma W, Jiang Z, Gu Y, Ma G, Ge H, Shen H, Lin Z. miR-422a inhibits glioma proliferation and invasion by targeting IGF1 and IGF1R. Oncol Res 2017; 25: 187-194.
- Lelli D, Pedone C, Sahebkar A. Curcumin and treatment of melanoma: The potential role of microRNAs. Biomed Pharmacother 2017; 88: 832-834.
- Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J, Ruan H, Ma S, Xu B. miR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. Onco Targets Ther 2017; 10: 1237-1246.
- Sun M, Wang X, Tu C, Wang S, Qu J, Xiao S. microRNA-216b inhibits cell proliferation and migration in human melanoma by targeting FOXM1 in vitro and in vivo. Cell Biol Int 2017.
- Villegas-Ruiz V, Hendlmeier F, Buentello-Volante B, Rodriguez-Loaiza JL, Miranda-Duarte A, Zenteno JC. Genome-wide mRNA analysis reveals a TUBD1 isoform profile as a potential biomarker for diabetic retinopathy development. Exp Eye Res 2017; 155: 99-106.
- de Oliveira Taveira M, Nabavi S, Wang Y, Tonellato P, Esteva FJ, Cantley LC, Wulf GM. Genomic characteristics of trastuzumab-resistant Her2-positive metastatic breast cancer. J Cancer Res Clin Oncol 2017; 143: 1255-1262.
- Loquai C, Schmidtmann I, Garzarolli M, Kaatz M, Kahler KC, Kurschat P, Meiss F, Micke O, Muecke R, Muenstedt K, Nashan D, Stein A, Stoll C, Dechent D, Huebner J. Interactions from complementary and alternative medicine in patients with melanoma. Melanoma Res 2017; 27: 238-242.
- Chen CF, Ruiz-Vega R, Vasudeva P, Espitia F, Krasieva TB, de Feraudy S, Tromberg BJ, Huang S, Garner CP, Wu J, Hoon DS, Ganesan AK. ATR mutations promote the growth of melanoma tumors by modulating the immune microenvironment. Cell Rep 2017; 18: 2331-2342.
- Babapoor S, Wu R, Kozubek J, Auidi D, Grant-Kels JM, Dadras SS. Identification of microRNAs associated with invasive and aggressive phenotype in cutaneous melanoma by next-generation sequencing. Lab Invest 2017; 97: 636-648.
- Alderman C, Yang Y. The anti-melanoma activity and oncogenic targets of hsa-miR-15a-5p. RNA Dis 2016; 3: 1450.
- Zhang D, Li Z, Zhang Y, Tu C, Huo J, Liu Y. miR-4262 promotes the proliferation of human cutaneous malignant melanoma cells through KLF6-mediated EGFR inactivation and p21 upregulation. Oncol Rep 2016; 36: 3657-3663.
- Qiu H, Yuan S, Lu X. miR-186 suppressed CYLD expression and promoted cell proliferation in human melanoma. Oncol Lett 2016; 12: 2301-2306.
- Hempel A, Kuhl SJ, Rothe M, Rao Tata P, Sirbu IO, Vainio SJ, Kuhl M. The CapZ interacting protein Rcsd1 is required for cardiogenesis downstream of Wnt11a in Xenopus laevis. Dev Biol 2017; 424: 28-39.
- Jannesari-Ladani F, Hossein G, Izadi-Mood N. Differential Wnt11 expression related to Wnt5a in high- and low-grade serous ovarian cancer: implications for migration, adhesion and survival. Asian Pac J Cancer Prev 2014; 15: 1489-1495.
- Liu D, Zhou K, Li Q, Deng F, Ma Y. Expression of Wnt11 and Rock2 protein with clinical characteristics of esophageal squamous cell carcinoma in Kazakh and Han patients. Int J Clin Exp Pathol 2015; 8: 7122-7130.
- Wei H, Wang N, Zhang Y, Wang S, Pang X, Zhang S. Wnt-11 overexpression promoting the invasion of cervical cancer cells. Tumour Biol 2016; 37: 11789-11798.