- Journal of RNA and Genomics (2006) Volume 2, Issue 1
In ovo RNAi opens new possibilities for temporal and spatial control of gene silencing during development of the vertebrate nervous system
| Thomas Baeriswyl and Esther T Stoeckli Institute of Zoology, University of Zurich, Winterthurerstrasse 190, 8057 Zurich, Switzerland |
| Correspondence to: Esther Stoeckli, Email: Esther.Stoeckli@zool.unizh.ch, Tel: +41 44 6354840, Fax: +41 44 6356879 |
| Journal of RNAi and Gene Silencing (2006), 2(1), 126-135 |
| © Copyright Baeriswyl and Stoeckli |
| (Received 01 December 2005; Revised 27 January 2006; Accepted 03 February 2006, Available online 28 February 2006; Published 28 February 2006) |
Abstract
Loss-of-function approaches are important tools for functional gene analysis. Due to the availability of sophisticated methods to manipulate gene expression in embryonic stem cells that can be used to generate mutant mice, the mouse is by far the most important vertebrate model organism for basic and applied biomedical research. Unfortunately, the available methods do not allow for precise temporal and spatial control of gene silencing during embryonic development limiting the usefulness of the mouse for developmental studies. Due to their easy accessibility chicken embryos have been one of the preferred model organisms for developmental studies. Their disadvantage, the lack of genetic tools, could be overcome by the development of in ovo RNAi (in ovo RNA interference), a method that allows for temporal and spatial control of gene silencing in vivo.
Keywords |
| RNAi, chicken embryo, in ovo RNAi, in utero RNAi, nervous system, embryonic development |
Introduction |
| Methods of gene silencing are important tools with applications from basic research to drug development and therapy. Different needs in these fields of applications have brought forth different solutions. Due to the possibilities to block gene function in large-scale screens invertebrates were the model organisms of choice for the molecular analysis of physiological and developmental processes (Friedman and Perrimon, 2004). In so-called forward genetic screens mutagens were used to randomly mutate genomes of flies and worms. The resulting phenotypes of interest were selected and the genes containing the mutation causing these phenotypes were identified. Unfortunately, the elegance of these forward genetic screens cannot be transferred to reverse genetics in these organisms. It is much more difficult to cause a mutation in a specific target gene and then look at the consequences of this mutation. However, reverse genetics is required to study the role of genes in a given context or to study gene function in vertebrates, where forward genetic screens are largely restricted to zebrafish. Although there are attempts to apply forward genetics to mice the high cost and the requirement for large space will keep their numbers low (Carlson and Largaespada, 2005; Kile and Hilton, 2005). Many questions, for instance in organogenesis or neurobiology, cannot be studied in invertebrates and require the analysis of gene function in vertebrates. The mouse has become the model organism of choice for the majority of questions in basic and biomedical research. Mice are easy to breed and house in a lab environment. For many aspects of human physiology and disease there would be a better model than the mouse but none of them offers a comparable toolkit for gene manipulations. Because mice can be reconstituted from embryonic stem (ES) cells that can be manipulated in culture sophisticated manipulations of the mouse genome are possible (for a recent review see Glaser et al, 2005). Still, there are limitations in using the mouse as a model when it comes to developmental studies. Due to the inaccessibility of mammals during gestation oviparous animals, i.e. fish, reptiles, and birds, are much easier to use for experimental manipulations in vivo. Their embryonic development is very similar and directly comparable to mammals, at least for those animals that do not undergo metamorphosis. However, the big disadvantage of fish, reptiles, and birds as model systems is the lack of genetic tools. RNAi is about to change that (Dykxhoorn and Lieberman, 2005). In fact, RNAi opens new possibilities of gene silencing in a temporally and spatially controlled manner that allows for studies that would be impossible with the available classical genetic tools. Spatial restriction can be achieved in mice by the use of sophisticated CreLox technologies and inducible promoters. These allow for the change in gene expression in the adult mouse without affecting gene function during development. However, there is still no way to control gene expression temporally in the precise manner that is required for studies of embryonic development. In this review we describe the advantages of RNAi technology for functional gene analysis during organogenesis using the nervous system as an example. We discuss the advantages and disadvantages of different model organisms in this context. Based on its easy accessibility during embryonic development the chicken is one of the preferred model organisms for developmental studies (Stern, 2005). The applicability of RNAi in chicken embryos made this model organism a perfect system to study gene function in a wide variety of tissues throughout development. |
Analysis Of Gene Function During Development Of The Nervous System |
| The development of an organism requires a precise timing of gene expression in a spatially restricted manner. Many genes play a role in different tissues and during more than one time window. Excellent examples demonstrating this are morphogens (Tabata and Takei, 2004, see below). Morphogens are signaling molecules involved in very early aspects of development. They act in a concentrationdependent manner on responsive cells to induce their differentiation to a particular cell type (Ashe and Briscoe, 2006). Morphogens include members of the hedgehog, the Wnt, the Fgf, and the TGFC family. More recently, new roles for morphogens during later stages of development have been discovered (Stoeckli, 2006; Charron and Tessier-Lavigne, 2005; Zou, 2004; Ciani and Salinas, 2003; Salinas, 2003). These studies have been possible thanks to sophisticated loss-of-function approaches in mice by restricting loss of gene function to a specific cell type (Charron et al, 2003) or by taking advantage of temporal control of gene silencing in the chicken embryo (Bourikas et al, 2005a). |
| Due to its complexity the nervous system takes a long time to develop. In fact in many species including humans neural development extends well beyond birth. It includes a variety of processes and genes that are involved in the development of other organs. For instance, genes involved in cell migration are often the same in the developing nervous system, in the heart or the vascular system (Carmeliet and Tessier-Lavigne, 2005). This has often hampered the analysis of genes by loss-of-function approaches in the nervous system, as mouse embryos died due to cardiac defects or defects in vasculogenesis and angiogenesis before their brains were fully developed. Alternatively, genes that have a function during several phases of embryonic development can only be studied during the earliest phase of activity, as their function during later stages is masked by aberrant initial development. Therefore, both classical reverse genetics as well as forward genetic screens have their limitations for the analysis of gene function during later stages of embryonic development. |
Model Organisms For Developmental Neuroscience |
| Although forward genetic approaches in invertebrate animal models like Drosophila melanogaster and Caenorhabditis elegans contributed much to our understanding of neural development, for many questions vertebrate model systems are required (Anderson and Ingham, 2003). Depending on a developmental neuroscientist’s demands, the vertebrate model organism of choice has to fulfil several criteria including availability of techniques for gene manipulation as well as easy accessibility during development. In addition, general requirements for the usefulness of a species as model organism have to be considered, such as the amount of money and time required for generating mutants, the availability in sufficient numbers and the easiness of husbandry of a model organism (Table 1). |
| The mouse: From conventional knockouts to in utero RNAi |
| The mouse is the most widely used model organism in developmental neuroscience, because techniques for lossand gain-of-function approaches based on homologous recombination in ES cells are available (Carlson and Largaespada, 2005; Kile and Hilton, 2005). However, creating knockout animals by using homologous recombination in ES cells is still very time-consuming and cost intensive, and therefore its usefulness for the analysis of a large number of genes is limited. Furthermore, conventional gene knockout strategies may result in embryonic lethality precluding the analysis of gene function in the developing nervous system. As mentioned above, disrupting the expression of a gene of interest early in development prevents any further functional analysis at later stages because cell types or entire structures may not form (Chiang et al, 1996; Ihle, 2000). |
| To overcome problems of conventional knockouts, recombinase systems under the control of cell or tissuespecific promoters have been developed to allow conditional gene knockouts in mice (Gawlik and Quaggin, 2004). In addition to the widely used CreLoxP system, two other recombinase systems have been used successfully: the Flp-FRT system from Saccharomyces cerevisiae and the phiC31 integrase (Dymecki, 1996; Belteki et al, 2003). Although conditional knockout technology using recombinase systems have provided insight into neural development, these strategies are limited by the requirement of cell- or tissue-specific promoters (Zhu et al, 2001; Blaess et al, 2004; Lewis et al, 2004). Furthermore, two transgenic mouse lines are required to knockout one gene: One mouse line that derives the Cre recombinase under the control of the tissue-specific promoter, and the other expressing the target gene flanked by loxP sites. Some temporal control of gene expression in adult mice has been achieved with the development of tetracyclinesensitive and tamoxifen-inducible Cre recombinase systems (Lewandoski, 2001; Metzger and Chambon, 2001; Morozov et al, 2003). However, these systems do not allow short-term switches of gene expression that are required during embryonic development. |
| Additional problems with knockout mice are genetic redundancy. Other members of the family of the targeted gene can compensate for the loss of a gene’s function to a degree that silencing one gene would not result in a detectable loss-of-function phenotype, hence requiring the generation of double- or triple knockout mice. The difficulties in generating conditional double or triple knockout mice would further complicate or prevent functional gene analysis during development. |
| RNA interference (RNAi), a conserved response to dsRNA resulting in specific gene silencing, represents an alternative way of blocking gene expression to conventional and conditional knockout technologies in mice (Lewis et al, 2002; McCaffrey et al, 2002; Prawitt et al, 2004). Hasuwa and colleagues showed long-term downregulation of EGFP in variety of organs of adult transgenic mice with a transgene-based RNAi system (Hasuwa et al, 2002). They used the polymerase III promoter H1 to drive expression of an shRNA. |
| Adenovirus-mediated RNAi resulting in specific gene silencing in mouse brain has been established and offers the possibility for temporal control of gene silencing in adult mice (Xia et al, 2002). Because mouse embryos develop in utero they are not easily accessible during prenatal stages for in vivo manipulations. RNAi in post-implantation mouse embryos using electroporation has been developed to knockdown genes during embryonic development (Calegari et al, 2002). Thus, very short-term experiments are possible because culture procedures for mouse embryos have been developed (Calegari et al, 2004). However, the time window for these mouse embryo culture systems is restricted to two days and available only for embryos between E7 and E13. Therefore the embryonic stages that can be studied are very limited. In order to study long-term functions of genes involved in brain development in utero electroporation guided by ultrasound has been developed for mice and rats (Takahashi et al, 2002; Bai et al, 2003). In contrast to whole embryo cultures, the embryos electroporated in utero can be maintained and analyzed from early embryonic to postnatal or adult stages. In utero electroporation in combination with RNAi has been used for the functional characterization of doublecortin during cortical development (Bai et al, 2003). Low efficiency and the requirement for expensive equipment for in utero electroporation limit the wide applicability of this approach, as does the problem of low spatial resolution. |
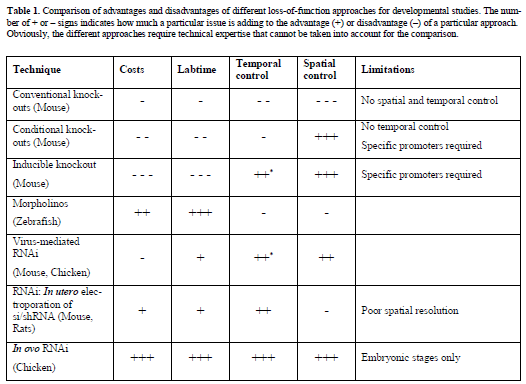 |
| *Temporal control is not available for embryonic stages |
| The Zebrafish as a model organism for developmental neuroscience |
| The zebrafish is a small tropical fish that represents an alternative vertebrate model organism to the mouse because of its rapid development ex utero. Embryos are translucent and therefore ideal for in vivo imaging. Improved methods for mutagenesis, transgenesis and gene targeting increase the usefulness of the zebrafish as a model organism for functional genomics (Ekker, 1999; Patton and Zon, 2001; Udvadia and Linney, 2003). Although chemical screens are highly effective in generating loss-of-function mutants, the process of identifying the mutated gene is laborious (Zhang et al, 1998; Talbot and Schier, 1999). Furthermore, as mentioned above, forward genetic approaches may not be useful for specific questions and do not allow for spatiotemporal control of gene silencing. |
| Antisense technology is a useful tool for specific gene silencing during development and has been applied in many species (Audic et al, 2001; Coonrod et al, 2001; Howard et al, 2001; Yang et al, 2001; Kos et al, 2003). Chemical modification of oligonucleotides has improved their stability and therefore increased their applicability in vivo. Morpholino phosphorodiamidate oligonucleotide-mediated gene inactivation is widely used for the analysis of gene function in zebrafish (Summerton and Weller, 1997; Nasevicius and Ekker, 2000; Corey and Abrams, 2001; Heasman, 2002; Sumanas and Larson, 2002). Morpholinos show a lower cellular toxicity and fewer side effects compared to conventional antisense nucleotides (Pickart et al, 2004). Usually morpholinos are microinjected into zebrafish embryos between the one- and the eight-cell stage. For effective gene inactivation the morpholino has to be complementary to the 5’UTR or the translation initiation site (Summerton and Weller, 1997; Heasman, 2002). The degree of gene silencing depends on the injected morpholino concentration and the extent of dilution due to cell proliferation (Heasman, 2002). Because morpholinos have to be injected into zebrafish embryos at very early developmental stages they loose effectiveness after a few days. Thus, functional analysis of genes expressed at later developmental stages cannot be achieved by this approach. The use of high concentrations of morpholinos increases the risk of inducing non-specific and toxic effects including cell death and neural degeneration (Nasevicius and Ekker, 2000; Braat et al, 2001; Karlen and Rebagliati, 2001; Lele et al, 2001). Lipofection can be used to improve cellular penetration of antisense oligonucleotides in vitro as well as in vivo (Juliano et al, 1999; Stenkamp et al, 2000). However, lipofection is associated with toxicity in vitro and even more importantly in vivo. |
| In ovo RNAi - a tool for functional gene analysis in chicken embryos allows for temporal control of gene silencing |
| For a long time the chicken embryo was a classical model organism for developmental studies in vertebrates because of its easy accessibility during development (Bourikas and Stoeckli, 2003; Bourikas et al, 2005b; Stern, 2005). In ovo as well as ex ovo culture methods of chicken embryos offer the possibility for in vivo manipulations throughout embryonic development (Perry, 1988; Stoeckli, 2003; Krull, 2004; Luo and Redies, 2005). After 21 days, at the time of hatching, the nervous system is fully developed and functional. The chicken genome is sequenced, and thus, comparisons of chicken genes with the human and the mouse or rat genomes are very easy (Hillier et al, 2004). |
| Due to biological constraints and the lack of ES cells it is not possible to manipulate the chicken genome with the same toolkit that is available for the mouse. The technology to generate transgenic chickens has been developed very recently (Mozdziak et al, 2003; Chapman et al, 2005), but the size and the long generation time of chickens limit the feasibility to breed them in a lab animal facility. |
| The easy accessibility of the chicken embryo during development of the nervous system was exploited for functional studies at the protein level using function-blocking antibodies (Stoeckli and Landmesser, 1995; Stoeckli et al, 1997; Burstyn-Cohen et al, 1999; Perrin et al, 2001). However, the limited availability of function-blocking antibodies severely restricted the usefulness of this approach. As an alternative, viral vector-based expression systems were developed to express dominant-negative proteins (Morgan and Fekete, 1996; Logan and Tabin, 1998). Morpholinos were also used successfully in chicken embryos (Kos et al, 2003; Tucker, 2004), although they may be more difficult to target to specific tissues than regular oligonucleotides (Krull, 2004). More recently, in ovo electroporation as an efficient method of gene transfer in chicken embryos for the temporally and spatially controlled ectopic expression of a gene of interest was established (Table 2; Muramatsu et al, 1997; Momose et al, 1999; Nakamura and Funahashi, 2001; reviewed in Bourikas and Stoeckli, 2003). Because loss-of-function phenotypes are usually more informative than gain-of-function phenotypes for the functional characterization of a gene of interest (Hudson et al, 2002) both viral vector-mediated and electroporation-based gene transfer depended on the availability of a dominant-negative mutant of the gene of interest. |
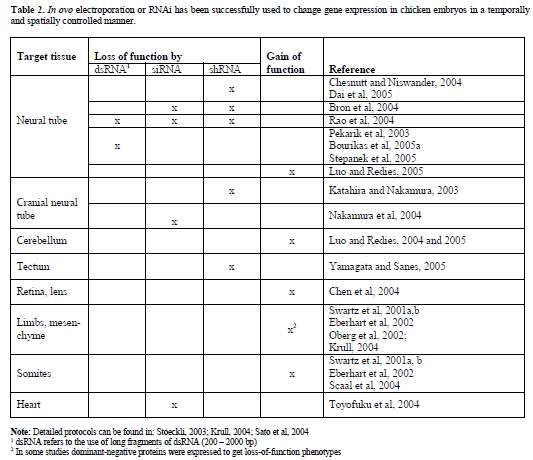
| To overcome the limitations of the chicken embryo as a model system for functional gene analysis, in ovo RNAi, a combination of in ovo electroporation for efficient nucleic acid transfer and RNAi for specific gene silencing has been established (Figure 1; Pekarik et al, 2003; Stoeckli, 2003; Bron et al, 2004; Krull, 2004). |
| Gene silencing has been achieved with several approaches and in many different areas of the nervous system (Table 2) but also other tissues (e.g. Toyofuku et al, 2004). The parameters for electroporation have to be adapted to the age of the embryo and the target tissue (Itasaki et al, 1999; Krull, 2004; Luo and Redies, 2004). More important than the electroporator (see Krull, 2004, for a comparison of different brands) is the choice of electrodes. For a focal transfer of nucleic acids a needle electrode is often placed directly into the tissue (Oberg et al, 2002; Luo and Redies, 2004). For a more widespread transfer a non-invasive method with wire or platelet electrodes is chosen (Dai et al, 2005; Nakamura et al, 2004; Toyofuku et al, 2004; Matsuda and Cepko, 2004). Some researchers found sonoporation more effective than electroporation for nucleic acid transfer into mesenchymal tissue (Ohta et al, 2003, but see Swartz et al, 2001a; Eberhart et al, 2004). |
 |
 |
| Figure 1. In ovo RNAi is an efficient method to silence genes in a temporally and spatially controlled manner in the developing neural tube. The chicken embryo can be accessed through a window cut into the eggshell. Phosphate-buffered saline containing long dsRNA, siRNA, or a plasmid encoding shRNA and 0.04% Trypan Blue is injected into the neural tube of the developing embryo with a glass pipette (A). In order to visualize the area where the injected RNA was taken up or as a control a plasmid encoding GFP can be co-injected. Wire electrodes are positioned parallel to the longitudinal axis of the embryo. Due to the negative charge of the nucleic acids cells toward the side of the anode are transfected. On average, we achieved 60% transfection efficiency in the targeted area of the neural tube of a 3-day-old embryo using 5 pulses of 25 Volts and of 50 msec duration (Pekarik et al, 2003). We use a one second inter-pulse interval. Depending on the position of the electrodes the target area can be selected. Positioning the electrodes dorsally will only target dorsal cell of the developing neural tube, whereas a more ventral position will result in transfected cells all along the dorso-ventral axis (as shown in B). To analyze the resulting phenotypes induced by knockdown of the target gene a variety of methods can be used. As an example we illustrate visualizing the trajectory of dorsolateral commissural neurons. These neurons extend their axons toward the floor plate, the structure that forms the ventral midline of the spinal cord. Axons cross the ventral midline before turning rostrally along the contralateral side of the floor plate (Bourikas et al, 2005a). The trajectory of these axons can be visualized by application of the lipophilic dye DiI to the cell bodies of commissural neurons (C). The comparison between control embryos, injected with a plasmid encoding GFP only or a control siRNA, with embryos injected with the target-specific dsRNA would reveal phenotypes in axon pathfinding. In the situation shown here, these would be caused by cell non-autonomous functions of the target gene as commissural axons from the side contralateral to the electroporated area are traced. For detailed protocols see references in Table 2. |
| In contrast to mammalian cell lines and non-embryonic tissue long dsRNA can be used in chicken embryos without induction of unspecific effects. No general inhibition of protein synthesis or induction of apoptosis has been observed (Pekarik et al, 2003; Chesnutt and Niswander, 2004). Similarly, no unspecific effects were seen in mouse oocytes (Stein et al, 2005) and cell lines derived from embryonic tissue (Billy et al, 2001). To avoid unspecific effects in postnatal mice and cell lines short interfering RNAs (siRNAs) can be used (Caplen et al, 2001; Elbashir et al, 2001). They have been successfully used in chicken embryos as well (Katahira and Nakamura, 2003; Sato et al, 2004). |
| As electroporation does not affect 100% of the cells in the target area (Pekarik et al, 2003; Luo and Redies, 2005) it may be important to identify those cells that did take up the siRNA or the dsRNA. In many cases coelectroporation of a plasmid encoding EGFP has been chosen and was sufficient (Pekarik et al, 2003; Nakamura et al, 2004). However, for an unequivocal identification of transfected cells the use of a vector-based approach has been developed. Driven by a polymerase III promoter short-hairpin RNAs (shRNAs) are produced in the cell. Because the plasmid contains an IRES site and also encodes EGFP all transfected cells are easily identified. The commonly used pol III promoters H1 and U6 were found to work well in chicken embryos (Katahira and Nakamura, 2003; Chesnutt and Niswander, 2004; Bron et al, 2004; Dai et al, 2005). |
| No matter whether long dsRNA, siRNAs, or shRNAs are used, the knockdown of target genes has been found to be efficient. Obviously, RNAi only prevents the synthesis of new protein. It cannot remove the pre-existing protein from a cell. Thus, for most effective gene silencing the injection and electroporation has to be carried out before the onset of gene expression. Gene silencing by RNAi was found to be long lasting in non-proliferating cells (from at least 9 days to 3 weeks; Sato et al, 2004; Omi et al, 2004). In cell culture or in tissues where cells proliferate the effect is diluted with successive cell divisions and usually decreases after 3-5 days. The use of a mixture of 3-5 different siRNAs is generally considered to be more effective than the use of a single siRNA. For that purpose mixtures of siRNAs can be generated in vitro from long dsRNAs by digestion with RNase ONE (Rao et al, 2004). The production of siRNAs in situ from vectors encoding shRNAs was found to extend the length of gene silencing compared to siRNAs (Bron et al, 2004). |
| As mentioned above, it is not necessary to generate the mixture of siRNAs in vitro before injection as long as embryonic tissue is used. Long dsRNA was always effective in gene silencing in our hands, presumably because they always give rise to a mixture of siRNAs that contains many effective ones. If siRNAs are designed with algorithms that are either freely available (Ui-Tei et al, 2004; Nakamura et al, 2004) or commercially used by companies selling siRNAs, it can still be frustrating to find effective ones. |
| Concerns about so-called off-target effects, i.e. the silencing of one or several non-target genes due to full or partial sequence homology with the siRNA have been raised (reviewed in Jackson and Linsley, 2004). Obviously such an event cannot be fully excluded but there are some rules to minimize the risk of off-target effects (Qiu et al, 2005). Firstly, it is of course essential to carry out proper BLAST analyses and to avoid sequences that are found in genes other than the target gene. Secondly, more than one (mixture of) siRNA or long dsRNA fragment should be used independently, as it is unlikely that they would have the same off-target effect, i.e. silence the same non-target genes. Thirdly, the concentration of the siRNA should be as low as possible. Generally, unspecific effects are not expected when concentrations are 20 nM or lower. When using long dsRNA we routinely get effective silencing with dsRNA concentrations in the range of 0.1-1 nM. |
| It is difficult to compare the efficiency of gene silencing between different RNAi approaches in the absence of systematic studies. The percentage of gene knockdown correlates with the concentration of siRNA, or dsRNA, respectively. Rao and colleagues compared the efficiency of siRNAs with a mixture of siRNAs produced in vitro from long dsRNA by RNase ONE, and long dsRNA (Rao et al, 2004). They concluded that siRNAs were more effective than the mixture of siRNAs created in vitro and long dsRNA, respectively. However, this conclusion is flawed by the fact that the number of effective siRNAs created from long dsRNA in vivo or in vitro by RNase ONE (where also ineffective siRNAs shorter than 21 bp are generated) is unknown. They used the same amount of siRNAs (200 ng/μl) and long dsRNA (700 bp fragment). Therefore, the concentration of the long dsRNA was roughly 30fold lower than the concentration of siRNAs. The effect of the much lower concentration of long dsRNA was still more than half as efficient as the siRNAs (53% compared to 90%), thus raising some doubts, whether siRNAs are really more efficient than long dsRNA. |
| The major advantages of in ovo RNAi are the temporal and spatial control of gene expression. This is due to the accessibility of the chicken embryo in ovo and the possibility to culture embryos ‘shell-less’ in dishes (Perry, 1988; Luo and Redies, 2005). Furthermore, it is easy to knockdown more than one gene at the same time. Fulllength cloning is not required, as cDNA fragments or ESTs can directly be used to produce dsRNA by in vitro transcription. Therefore, in ovo RNAi represents a fast and inexpensive tool for functional genomics. It can easily be adapted for the assessment of different developmental processes (Table 2; Bourikas and Stoeckli, 2003; Krull, 2004; Eberhart et al, 2004; Toyofuku et al, 2004; Luo and Redies, 2004 and 2005; Luo et al, 2004). |
| Taking advantage of the major asset of in ovo RNAi, i.e. precise temporal control over gene silencing during embryonic development, a role for the morphogen sonic hedgehog (SHH) in postcommissural axon guidance could recently be demonstrated (Bourikas et al, 2005a). During early stages of embryonic development SHH is involved in inductive and patterning processes including control of left-right asymmetry and formation of the limb (reviewed by Marti and Bovolenta, 2002; Jacob and Briscoe, 2003). Slightly later in development, Shh was shown to act in parallel to netrin-1 as a chemoattractant for dorsal commissural axons (Charron et al, 2003). All these effects of Shh are mediated by a receptor complex composed of Patched and Smoothened. Interestingly, these receptors are not involved in Shh’s effect on postcommissural axons. After crossing the floor plate, the ventral midline of the spinal cord, commissural axons are no longer expressing Patched and Smoothened. The repulsive effect of Shh on postcommissural axons is mediated by Hip (Hedgehog interacting protein). Thus, commissural axons switch from being attracted by Shh before midline crossing to being repelled by a concomitant switch in receptor expression (Figure 2; Bourikas et al, 2005a; reviewed by Charron and Tessier-Lavigne, 2005). The analysis of this rapid change in responsiveness to Shh would not have been possible without a method that allows for precise temporal control of gene silencing. |
| The large number of papers describing various approaches of RNAi-based gene silencing in chicken embryos demonstrates the versatility of the chicken embryo on the one hand and RNAi on the other hand. We have only been able to include some of the studies carried out in the last 2 to 3 years and focused largely on the development of the nervous system. The multitude of approaches, siRNAs versus shRNAs or long dsRNA, different electrodes used with different electroporation parameters may be confusing at first glance. However, transfection of a plasmid encoding GFP is an easy way to get started. It allows for fast assessment of transfection efficiency in the target area and for selection of experimental parameters. Above all, it provides a fast method to test and train the skills of the experimenter to handle chicken embryos in ovo or ex ovo. Beginners should use GFP expression to assess their skills with respect to reproducibility of electroporation and the absence of artefacts due to tissue damage caused by injection or by touching embryonic tissue with the electrodes. The fact that both in ovo RNAi and ex ovo RNAi require some manual skills for handling live embryos may in fact represent their biggest disadvantage. The best way to learn handling chicken embryos is by visiting a lab where they are routinely used for research. |
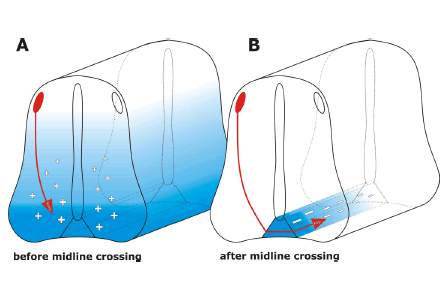 |
| Figure 2: In ovo RNAi allows for temporal control of gene silencing in the developing neural tube. In order to study gene function during embryonic development precise temporal and spatial control of gene silencing is required. Classical knock-out strategies do not allow for functional gene analysis during later stages of development, as lack of target gene expression during the first time window would preclude the analysis of its function during later stages. An example illustrating the requirement for temporal control is the analysis of SONIC HEDGEHOG (SHH). Shh is a morphogen that is required for differentiation of cells in the spinal cord during early stages of development (Jessell, 2000). Slightly later, Shh acts in parallel to Netrin-1 as a long-range guidance cue, attracting dorsal commissural axons toward the floor plate (A; Charron et al, 2003). This attractive effect of Shh is mediated by the co-receptor formed by Patched and Smoothened (Smo). A few hours later, after commissural axons have crossed the floor plate, Shh acts as a repulsive guidance cue, directing post-commissural axons rostrally (B; Bourikas et al, 2005a). The repulsive activity of Shh is mediated by Hedgehog-interacting protein (Hip). Thus, within a short period of time, commissural axons switch receptors (from Smo to Hip) that allow them to respond differently to Shh gradients. |
Conclusions |
| The development of in ovo RNAi has not only reinstated the importance of the chicken embryo as a model organism for developmental studies but it has added precise temporal control of gene silencing to our toolkit for gene manipulations. As demonstrated in recent studies, temporal and spatial control of gene function is an important aspect of functional gene analysis during embryonic development. |
Acknowledgements |
| Research in the lab of ET Stoeckli is supported by the Swiss National Science Foundation and the NCCR “Brain Plasticity and Repair”. |
Statement Of Compteting Interests |
| The authors declared no competing interests. |
References
|