- Journal of RNA and Genomics (2010) Research Article
Identification of small interfering RNA targeting Signal Transducer and Activator of Transcription 6: Characterisation and selection of candidates for pre-clinical development
| Gareth D Healey1,*, Shawn Zinnen1, Jennifer A Lockridge1,3, Ivan Richards1, Neil Evans1 and William Walker1,2 1Allerna Therapeutics Ltd, Institute of Life Science, Swansea University, Swansea, SA2 8PP, UK, 2School of Medicine, Institute of Life Science, Swansea University, Swansea, SA2 8PP, UK, 3Lockridge Pharmaceutical Consulting, LLC, 9689 Otis Drive, Westminster, CO 80021, USA |
| *Correspondence to: Gareth Healey, Email: ghealey@allerna.com, Tel: +44 (0) 1792602903 |
| Received 28 June 2010; Revised 18 August 2010; Accepted 20 August 2010; Published online 25 August 2010 |
| J RNAi Gene Silencing, 2010, 6(2), 401-410 |
| © Copyright The Authors: This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/2.0/uk/). This license permits noncommercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details. |
Abstract
The interleukin (IL)-13 pathway and its associated transcription factor, signal transducer and activator of transcription 6 (STAT6), have been clearly implicated in the pathogenesis of bronchial asthma. We have developed a system to effectively screen the STAT6 gene for targeting with small interfering (si) RNA molecules. By incorporating an in silico and in vitro screening system we were able to identify fourteen siRNA molecules suitable for pre-clinical drug development. Furthermore, we were able to demonstrate that modification of certain siRNAs, designed to improve in vivo longevity, was possible without significant loss of target knockdown efficacy and that the siRNA produced by our selection process did not induce demonstrable interferon responses. These data suggest that several STAT6-targeting siRNA suitable for preclinical development are available for potential use in the treatment of asthma.
Keywords |
| Asthma, STAT6, IL-13, siRNA, pre-clinical development |
Introduction |
| The recently identified roles of the interleukin (IL)-13 pathway (Huang et al, 1995; Wills-Karp et al, 1998; Zhu et al, 1999; Wills-Karp, 2001; Brightling et al, 2003), the Signal Transducer and Activator of Transcription 6 (STAT6) (Akimoto et al, 1998; Yang et al, 2001; Kuperman et al, 2002) protein and epithelial cells (Holgate 2007; Kato and Schleimer 2007) in the pathogenesis of bronchial asthma, have highlighted this system as a potential therapeutic target for asthma (Izuhara et al, 1999; Schaefer et al, 2001; Izuhara et al, 2006). While the current therapeutic approaches rely on the use of steroidbased therapies (Adams et al, 2007 Adams et al, 2008a; Adams et al, 2008b), our more targeted approach is to ablate the inflammatory responses caused by local expression of T helper type 2 (Th2) cytokines, which have been demonstrated to be associated with the allergic asthma phenotype (Larche et al 2003). In particular, IL-13 and its associated pathway have been shown to be a key component of the asthmatic response, which includes eosinophilic infiltration, mucus hyper-secretion, subepithelial fibrosis and airway hyper reactivity (Zhu et al, 1999). By targeting a key transcription factor (STAT6), which is unique to this pathway, we are able to stem the ongoing inflammatory response initiated by epithelial cell exposure to IL-13. |
| We have previously demonstrated the use of siRNA to ablate STAT6 messenger RNA and protein (Walker et al, 2009), and demonstrated that this was an effective approach to rapidly achieve attenuation of ongoing IL-13 mediated events in lung epithelial cells (Walker et al, 2009). |
| Here, we expand this work and demonstrate the screening of the entire STAT6 gene to identify suitable siRNA available for potential therapeutic development. We then show that chemical modification, to prevent degradation in vivo, can be achieved without detectable loss of efficacy and identify lead candidates for future pre-clinical development. |
Materials and Methods |
| Cell cultures |
| The human A549 lung epithelial cell line was obtained from European Collection of Cell Cultures (ECACC, Salisbury, UK) and routinely cultured in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) containing 10% (v/v) heat inactivated fetal bovine serum (HIFBS) (Invitrogen, UK) and 1% (w/v) penicillin/ streptomycin solution (Sigma-Aldrich, UK). Subconfluent cultures were split using trypsin/ethylenediaminetetraacetic acid (EDTA) (Invitrogen, UK). |
| Transfection |
| For transfection with siRNAs, 4x104 cells were seeded into each well of a 12-well tissue culture tray (Greiner Bio-One, UK) in 1ml DMEM containing 10% (v/v) HIFBS without antibiotics (unless otherwise stated). Cells were cultured at 37°C in a humidified 5% (v/v) CO2 environment for 24hr prior to transfection. siRNAs were then diluted to the desired concentration in OptiMEM serum-free medium (Invitrogen, UK) and transfections were performed using the chemical transfection agent Interferin™ (Poly Plus-Transfection, Inc) in accordance with the manufacturer’s instructions. Following transfection, cells were incubated at 37°C in a humidified 5% (v/v) CO2 environment for 72hr before sample collection. In certain experiments cells were cultured in the presence of recombinant human IL-13 (PeproTec EC, UK) at a final concentration of 50ng/ml for the last 24hr of the culture, or Poly (I:C) (Sigma-Aldrich, UK) at the final concentrations indicated. For experiments to test for interferon responses, cells were only cultured for 24hr following transfection before sample collection. |
| siRNAs |
| Unmodified siRNAs were purchased from Ambion Inc (Austin, TX), whilst modified siRNA were synthesised as single strands by Integrated DNA Technologies Inc, (Belgium). Annealing of the siRNA single strands was performed by mixing equal molar concentrations of each strand, heating to 90°C and then allowing cooling to room temperature. Confirmation of successful annealing was achieved on 15% (w/v) TBE-Urea polyacrylamide gels followed by visualisation with ethidium bromide (Sigma- Aldrich, UK) under UV light using a gel doc machine (Bio-Rad, UK). Scrambled control siRNA was Silencer® Negative Control #1 (Ambion, UK). |
| siRNA modification |
| In order to improve the in vivo stability, chemical modification of the siRNA was performed. The following chemical modifications were included in the siRNA design: |
| • Deoxyribonucleotide Adenosine, Uracil, Guanine and Cytosine (A, U, G, C) |
| • 2’ O-Methyl modified Adenosine or Guanine (mA, mG) |
| • Internal 2’ Fluoro modified Cytosine and Uracil (i2FC, i2FU) |
| • Phosphorothioate linkages (*) |
| Gene expression analysis |
| Total RNA was isolated from Tri-Reagent® (Sigma- Aldrich, UK) and first strand cDNA synthesis carried out from 1μg RNA using the High Capacity cDNA synthesis kit (Applied Biosystems, UK). Quantitative and semiquantitative real-time PCR was performed in a duplex reaction. This used TaqMan gene expression assays with a FAM probe (Applied Biosystems, UK) to the target gene and the TaqMan control assay (Applied Biosystems, UK) to the house keeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with a VIC-TAMRA probe. PCR master mix used was the TaqMan universal PCR master mix, no Amperase UNG (Applied Biosystems, UK) and reaction ratios were as per the manufacturer’s instructions. |
| Standard PCR conditions were 95°C for 10min, followed by 40 cycles of 95°C for 15sec and 60°C for 60sec with fluorescence quantification after each anneal/extension step. The expression of each gene was normalised against the expression of the house keeping gene GAPDH. For STAT6 analysis, quantification was performed using a standard curve of recombinant human STAT6 and results expressed as a percentage of total STAT6 remaining compared to a transfection reagent only (negative) control. For other genes, gene expression was compared to relative expression in the presence of transfection reagent only using the method described by Pfaffl (2001). |
| Measurement of protein expression by western blotting and enzyme-linked immunosorbent assay (ELISA) |
| Western blotting for STAT6 and GAPDH was performed, as previously described (Walker et al, 2009). Protein densitometry analysis was carried out using the Bio-Rad Quantity OneTM software. ELISA for human CCL26 (Eotaxin 3) was performed as previously described (Walker et al, 2009). |
| THP-1 interferon assay |
| The human THP-1 monocyte cell line was obtained from European Collection of Cell Cultures (ECACC, Salisbury, UK) and routinely cultured in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% (v/v) heat inactivated fetal bovine serum (HIFBS) (Invitrogen, UK) and 1% (w/v) penicillin/streptomycin solution (Sigma- Aldrich, UK) in a suspension culture. |
| For transfection, cells were removed from culture, washed and seeded into 6-well tissue culture trays (Greiner Bio- One, UK) in 2ml RPMI-1640 containing 10% (v/v) HIFBS without antibiotics. 12-O-tetradecanoylphorbol-13-acetate (TPA) was then added to each well of the trays at a final concentration of 200ng/ml and cells cultured at 37°C in a humidified 5% (v/v) CO2 environment for 72hr to induce maturation to a macrophage phenotype, which causes them to stop dividing and adhere to the well plastic. After 72hr, the medium containing TPA was removed from the wells and 2ml fresh RPMI-1640 plus 10% (v/v) HIFBS added. Transfection was then carried out as described above in ‘Transfection’. |
| Statistical and Data analysis |
| Data from multiple experiments are expressed as mean ± standard deviation. For mRNA fold-change data, differences between groups were examined for statistical significance using the Mann–Whitney U-test. Quantitative measurements were compared using Student’s t-test. In all cases, p values <0.05 were considered significant. |
| IC50 values were determined using the IDBS™ XLfit Excel add-in analysis package. |
Results |
| Design of siRNAs for the STAT6 gene |
| Initial bioinformatic screening of the STAT6 gene using NCBI BLAST® was carried out to select possible candidates for in vitro screening. This process involved breaking down the human STAT6 nucleotide sequence (GeneBank Seq ID: NM_003153) into all possible 19- mers, which were then aligned against the target sequences for mouse, rat and non-human primate (Rhesus) STAT6 (GeneBank Seq ID’s: NM_009284.2, NM_001044250 and XM_001115590.1, respectively). Only those sequences, which aligned with all four species, were carried forward. |
| A hierarchy of the 19-mers was then constructed. 19-mers with perfect sequence identity across all four of the target species were placed at the top followed by 19-mers with increasing degeneracy. Degeneracy was defined as follows: Wobble pairs were considered less of a problem then mis-pairs and gaps were considered the most degenerate. Any 19-mers with a run of four or more guanine bases was pushed down the list due to the manufacturing challenges inherent to such sequences, which are known to form polymorphic structures (Dai et al, 2007; Zhang et al, 2010). |
| Sequences were then blasted against the whole GenBank database using NCBI BLAST® to identify potential offtarget silencing. Identities of 16 or more bases were noted and if the gene affected was of concern, the 19-mer was moved down the list. Finally, a visual inspection of the sequences was carried out to finalise the hierarchy. Tighter base pairing at the 3’-end of the antisense relative to the 5’-end can be a benefit (Ui-Tei et al, 2004; Rana, 2007) and therefore was used as a criterion to move a 19- mer up the hierarchy. Purines and most favourably adenosines at the 5’-end of the anti-sense strand were also preferentially selected for to favour stabilisation chemistry if required (Ui-Tei et al, 2004; Rana, 2007). |
| This selection process left forty siRNA candidates to be taken forward for in vitro screening, which were designated the numbers 1 to 40. |
| Characterisation of STAT6 siRNA activity |
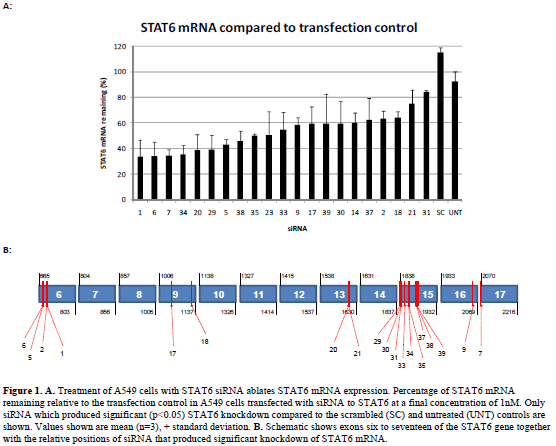
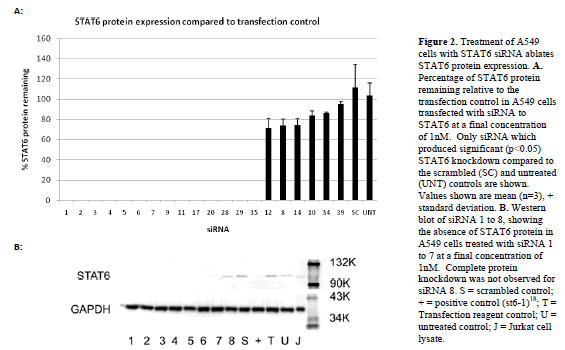
| Forty siRNAs were screened for inhibitory activity. Their ability to inhibit STAT6 expression at the mRNA and protein level was assessed using a quantitative (q) polymerase chain reaction (PCR) and western blotting, respectively. STAT6 mRNA in A549 cells was analysed after transfection with each of the siRNA at a final concentration of 1nM. Figure 1 shows STAT6 mRNA remaining (relative to the transfection control) for those siRNA, which produced significant (p<0.05) knockdown compared to the scrambled and untreated controls (21 out of the 40). The relative binding positions of each of these siRNAs within the STAT6 gene are illustrated in the schematic in Figure 1, demonstrating localisation to exons 6, 9, 13, 15, 16 and 17. siRNAs that did not produce significant STAT6 knockdown (not shown) were also found to localise to exons 6, 9, 13, 15 and 16. Western blotting was then performed to determine the degree of STAT6 protein knockdown following siRNA treatment at 1nM. Figure 2 shows a representative blot demonstrating the presence or absence of STAT6 protein for siRNAs 1-8. The relative STAT6 band densities for those siRNAs, which produced significant (p<0.05) knockdown compared to the scrambled and untreated controls (20 out of 40), is also shown (Figure 2). |
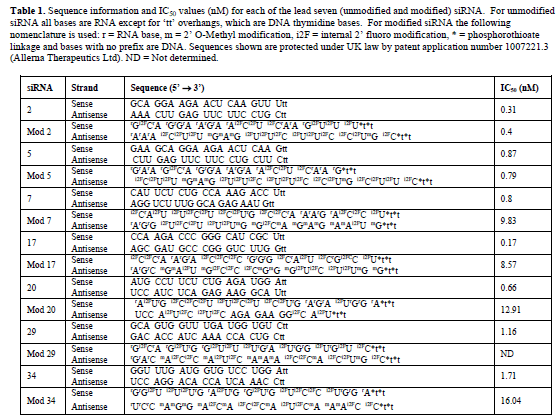
| Candidates for the next phase of testing were selected on the basis of significant STAT6 mRNA and protein knockdown, which left fourteen siRNA that were subjected to a second NCBI BLAST® analysis. This identified theoretical mis-match events and/or potential off-target effects in at least one of the four target species, in seven of the siRNAs. As a result these seven siRNAs were designated as backup development candidates, leaving siRNAs 2, 5, 7, 17, 20, 29 and 34 for further development (siRNA sequence information is shown in Table 1). |
| Determination of the 50% inhibitory concentration (IC50) for unmodified siRNA |
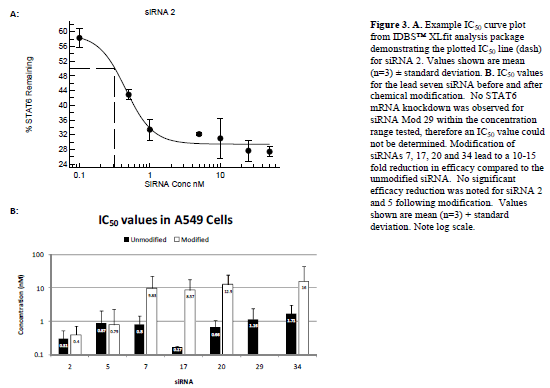
| IC50 values for each of the seven lead siRNA (Figure 3 and Table 1) were next determined by repeating the transfection experiments with a range of siRNA concentrations from 0.1nM to 100nM. Results demonstrated that all seven of the siRNAs were highly efficacious with IC50 values in A549 cells of less than 2nM in all cases and less than 1nM for siRNAs 2, 5, 7, 17 and 20. |
| Determination of the 50% inhibitory concentration (IC50) for chemically modified siRNA |
| IC50 experiments were then repeated for modified versions of each of the siRNA (Figure 3 and Table 1). Modification of siRNAs 7, 17, 20 and 34 led to a 10-15 fold increase in IC50 value in A549 cells, whilst the reduction in efficacy for siRNA 29 following modification was so pronounced (> 100-fold) that it was not possible to determine an IC50 within the concentration range of the experiment (0.1 to 100nM). No significant reduction in efficacy was noted for siRNAs 2 and 5 following modification; therefore, siRNAs 2 and 5, with and without modification were taken forward for further development. |
 |
 |
 |
 |
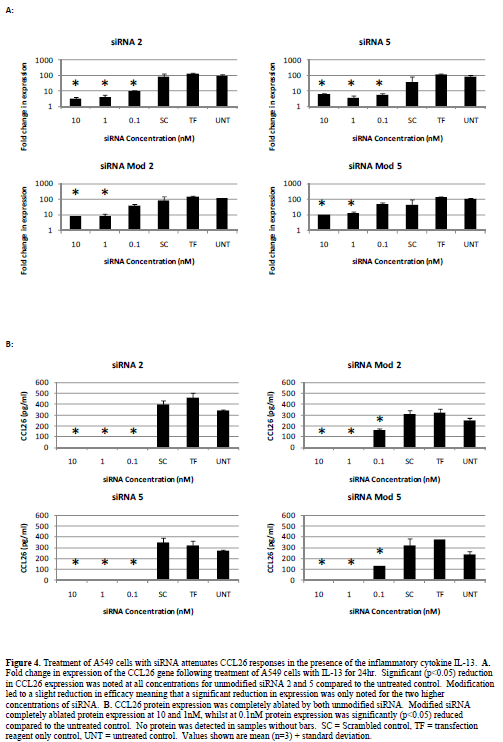
| Attenuation of Eotaxin 3 (CCL26) responses |
| We previously demonstrated that STAT6 siRNA potently suppress ongoing inflammatory events in the presence of inflammatory cytokines (Walker et al, 2009). We also demonstrated that CCL26 (eotaxin 3) is the predominant eotaxin chemokine up-regulated in A549 cells in response to the inflammatory cytokine IL-13 and that there is no significant difference between IL-13 and IL-4 in terms of ability to up-regulate eotaxin mRNA expression (Walker et al, 2009). We therefore utilised the up-regulation of CCL26 mRNA and protein as a functional downstream test of the robustness of STAT6 ablation in our model system. A549 cells treated with IL-13 (50ng/ml) for 24hr show a significant up-regulation in CCL26 gene and protein expression (Figure 4). Treatment with unmodified siRNA 2 or 5 significantly (p<0.05) reduced CCL26 gene expression and completely ablated protein expression at all concentrations tested. Modification of siRNAs 2 and 5 demonstrated approximately a 10-fold reduction in efficacy, with the higher concentrations tested still showing attenuation of CCL26 gene expression and complete loss of protein expression. No reduction in gene expression was noted following treatment with modified siRNA at 0.1nM although a significant reduction in protein expression compared to the untreated control was still observed. |
| Interferon responses |
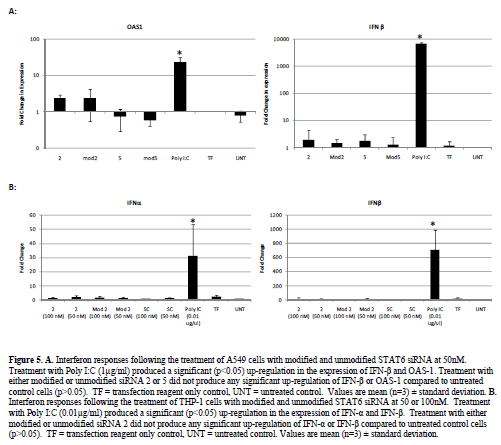
| The presence of interferon responses associated with long double stranded RNA has been reported in previous siRNA studies (Bridge et al, 2003; Sledz et al, 2003). We therefore, carried out screening by RT-PCR for interferon (IFN)-α, IFN-β and the interferon induced gene 2’5’-oligoadenylate synthase (OAS)-1. No significant up-regulation of either IFN-β or OAS-1 was noted following the transfection of A549 cells with either modified or unmodified siRNA (50nM) compared to untreated control cells. Treatment with Poly I:C (1μg/ml) however did produce a significant (p<0.05) up-regulation of both genes (Figure 5). |
| We then further tested one of our siRNA sequences in a THP-1 cell based interferon assay, which we had found to be more sensitive to stimulation with double stranded RNA, requiring only 0.01μg/ml of Poly I:C to generate significant up-regulation of the IFN–α and IFN-β genes. Stimulation of THP-1 cells with either the modified or unmodified siRNA 2 molecule (at 50 or 100nM) produced no significant (p>0.05) up-regulation of the IFN-α and IFN-β genes compared to untreated control cells (Figure 5). Treatment with Poly I:C (0.01μg/ml) however did produce a significant (p<0.05) up-regulation of both genes (Figure 5). |
| Despite previous studies suggesting the presence of interferon responses to siRNA, these data corroborate our previous work (Walker et al, 2009), which demonstrated that our STAT6 siRNA do not induce an interferon response. |
Discussion |
| The complexity of the asthma phenotype and complications associated with current treatment regimens point to the need for an effective, side-effect-free approach to the treatment of asthma. Several groups have demonstrated that the IL-13 pathway is a key mediator of airway hyperreactivity, mucus production and eosinophilia within the asthmatic lung (Wills-Karp et al, 1998; Wills-Karp, 2001; Yang et al, 2001; Kuperman et al, 2002; Izuhara et al, 2006; Bree et al, 2007). Central to this signalling pathway is STAT6, the importance of which has been demonstrated with gene-knockout mice (Akimoto et al, 1998; Kuperman et al, 2002) and in studies of human disease (Christodoulopoulos et al, 2001; Mullings et al, 2001). We previously showed that an RNAi-based strategy offers superior targeting of STAT6 gene expression within the A549 lung epithelial cell-line (Walker et al, 2009). We also demonstrated the suitability of this model through similarity of IL-13 responsiveness and STAT6 knockdown by RNAi with cultured primary small airway epithelial cells and bronchial smooth muscle cells (Walker et al, 2009). Thus having previously designed efficacious STAT6 siRNA, we sought to derive all STAT6 siRNA suitable for pre-clinical in vivo testing and explore chemical modification with a view to facilitate in vivo studies. |
| We therefore developed a rapid screening capability through a combination of in silico and in vitro approaches, which enabled us to determine those siRNA suitable for further in vivo characterisation. Pre-clinical characterisation of siRNA requires the use of animal models for efficacy and toxicity studies. Thus, in addition to the human STAT6 sequence we utilised GeneBank STAT6 sequences for three species likely to be used as part of a pre-clinical development programme, mouse, rat and nonhuman primate. By aligning potential siRNA sequences against STAT6 sequences from all four species and then ranking them within a hierarchy based on sequence and manufacturing suitability, we were able to significantly reduce the size of our siRNA pool. This served to facilitate a more rapid in vitro screening process and also ensured we avoided siRNA that may require complicated and expensive manufacturing processes. The suitability of this approach is exemplified by the fact that our in silico process delivered fourteen highly efficacious siRNA, which when transfected into A549 cells at 1nM, produced significant (p<0.05) STAT6 mRNA and protein knockdown compared to scrambled and untreated controls. Further characterisation of our lead candidates demonstrated that all exhibited highly efficacious IC50 values between 0.1 and 2nM suggesting all would be suitable for in vivo characterisation. However, despite studies demonstrating successful RNAi-mediated gene targeting within mouse lung tissues (Massaro et al, 2004; Bitko et al, 2005; Thomas et al, 2007; Darcan-Nicolaisen et al, 2009), we considered chemical modification of our siRNA as a means to improve in vivo stability and therefore increase the likely efficacy of candidates taken forward for in vivo development. The rationale for modification was predicated on the assumption of benefit if both strands possess increased resistance to degradation. While this concept has not yet been tested, we assumed the more critical anti-sense strand should contain the greatest stabilisation and therefore allowed an imbalance favouring degradation of the sense strand after RISC loaded onto the target site. Thus, all sense strands were modified with 2’- fluoropyrimidines whilst the A and G purines remained natural ribose; the antisense strands were reinforced with additional 2’-Ome modification of their purines. Furthermore, we strove to maintain the critical 5’-terminus of the antisense strand in the least modified form possible. When the antisense strand contained purines at the three 5’ positions they remained as ribose. If any of the three 5’ positions included a pyrimidine it was modified due to its more nuclease sensitive nature. Typically, chemical modification of siRNA is associated with a loss in efficacy together with improved pharmacokinetics and reduced immune stimulation (Morrissey et al, 2005; Rana, 2007). We therefore subjected chemically modified forms of our siRNA to further IC50 analysis to determine any efficacy loss associated with modification. Whilst five of our seven leads demonstrated losses in efficacy consistent with the literature (Morrissey et al, 2005; Rana, 2007), we also noted two candidates that showed no significant (p>0.05) reduction in IC50 value (in terms of STAT6 mRNA knockdown) compared to their unmodified forms within our A549 model. It is also acknowledged that an exhaustive analysis of chemically modified sequences is not possible. Therefore, we cannot exclude the possibility of siRNAs with both increased activity and stability. In particular, it is noted that the use of 3’dTdT overhangs has been shown to reduce the duration of action of siRNA both in vitro and in vivo (Hohjoh, 2002; Strapps et al, 2010). We will therefore look to testing alternative 3’-overhang chemistry in the future studies. |
 |
 |
| The allergic asthmatic lung is characterised by the infiltration of eosinophils into peribronchial sites, which occurs under the control of IL-13. Secretion of IL-13 by lymphocytes within the lung leads to local production of eotaxin chemokines, which act as chemo-attractants recruiting eosinophils to the airways where subsequent degranulation leads to airway damage and remodelling (Davies et al, 2003). The importance of the eotaxin family of chemokines to the asthma phenotype and their association with airway remodelling and fibrosis (Joubert et al, 2005; Beck et al, 2006), led us to suggest that blockade of the eotaxin pathways will likely be an essential component of future asthma therapeutics. We subsequently demonstrated that RNAi of STAT6 prevented IL-13-mediated eotaxin family mRNA induction and ablated functional CCL26 protein expression (Walker et al, 2009). Within the current study we have again demonstrated that our lead siRNA candidates, both modified and unmodified, potently ablate both CCL26 mRNA and protein expression in the presence of IL-13. CCL26 analysis however, did provide a differentiator between modified and unmodified siRNA. Whilst at the STAT6 mRNA level no significant difference was noted between the modified and unmodified versions of our lead siRNA, CCL26 ablation did show a difference. At the lowest concentration tested (0.1nM), neither modified siRNA 2 or 5 produced significant knockdown of CCL26 mRNA or protein, whilst the unmodified forms did. Whilst this finding suggests reduced efficacy following modification corroborating findings within the literature, we have still shown that chemical modification of siRNAs to achieve in vivo stability is possible with only minimal loss of efficacy. Furthermore, work to characterise these siRNAs in vivo is required to demonstrate the balance of improved longevity in vivo against target knockdown efficacy. |
| In addition to improved in vivo longevity, another important facet of the chemical modification of siRNAs is to prevent the induction of interferon responses, which have been reported in the literature (Bridge et al, 2003; Sledz et al, 2003). Within the current study we have demonstrated an absence of interferon activation in two human cell culture systems suggesting that our siRNA molecules do not induce such a response. In addition to toll-like receptor (TLR) 3 mediated immune stimulation (already investigated herein), several groups also report the immunostimulatory effects of siRNA via TLR 7 and 8 (Hornung et al, 2005; Judge et al, 2005). Whilst it will clearly be important to test for such an effect in future in vivo work, within the current study such investigation is not appropriate for a number of reasons. As a potential asthma therapeutic, the predominant cell type targeted by our siRNA will be airway epithelial cells, which express TLR 3 but not TLR 7 or 8. Whilst dendritic cells will undoubtedly be encountered within the lung, the appropriateness of a peripheral blood mononuclear cell (PBMC) model is diminished since we are not considering intravenous (iv) as a potential route of administration, rather inhalation. It has been demonstrated that the administration of naked siRNAs (without any delivery or transfection agent) fails to generate any interferon responses (Hornung et al, 2005; Judge et al, 2005). Our own work also corroborates this finding, whereby the administration of Poly IC to our cell models without a transfection reagent failed to stimulate the up-regulation of interferon α/β mRNA (data not shown). As a result, work demonstrating TLR 7/8 mediated responses incorporated the use of microencapsulation to facilitate siRNA uptake. This coupled with the iv administration route chosen, would have specifically targeted the TLR 7/8 rich endosomal compartment of dendritic cells, providing an opportunity for TLR 7/8 stimulation. Although, our use of the PEI based transfection reagent Interferin™ would also target this compartment, such reagents have been shown to facilitate quick exit from the endosome by inducing rapid proton influx and endosomal swelling and rupture (Akinc et al, 2005). Such a mechanism would significantly reduce the time available for endosomal degradation and also likely negate the possibility of endosomally localised TLR stimulation. As a consequence, we chose to only employ TLR 3 stimlation as a factor within our in vitro screening system. Further in vivo testing however, will enable us to determine whether the non-stimulatory nature of our siRNA is only true within an in vitro culture system and our modifications may then accrue further benefit once in vivo. |
| Several studies have now been published demonstrating the use of RNAi to target epithelial cell STAT6 both in vitro and in vivo (Rippman et al, 2005; Darcan-Nicolaisen et al, 2009; Walker et al, 2009). Other studies however, have used high concentrations of siRNA and failed to show complete inhibition of expression of STAT6 and/or downstream signalling events. As previously, our extensive dose response analyses have demonstrated that effective inhibition of expression of STAT6 and downstream signalling events can be achieved with low concentrations of non-interferon-inducing STAT6 targeting siRNA. Furthermore, we have shown that it is possible to achieve knockdown at these concentrations with siRNA chemically modified to improve in vivo stability and therefore increase the likely in vivo efficacy. These data support the suitability of STAT6 as an important and viable target for the therapy of asthma and further work within our lab will test the therapeutic potential of STAT6 siRNA within the asthmatic model. |
Conclusions |
| The IL-13 pathway and its associated transcription factor, STAT6, have been clearly implicated in the pathogenesis of bronchial asthma. |
| Identification of efficacious siRNA to STAT6 was achieved through the use of a combined in silico and in vitro screening approach. |
| Chemical modification strategy achieved without significant loss of in vitro efficacy. |
| Several STAT6-targetting siRNA suitable for preclinical development were identified. Further work will investigate in vivo efficacy. |
Acknowledgements |
| The work described herein was wholly funded by Allerna Therapeutics Ltd. |
Compteting Interests |
| The authors declare that there are no significant financial, professional or personal competing interests that may have influenced the performance or presentation of the work described in the manuscript. Gareth Healey and Neil Evans are employees of Allerna Therapeutics Ltd, all other authors are employed as consultants to Allerna Therapeutics Ltd. Sequences disclosed are protected by US patent # US 7,566,700 B2 and patent application # 1007221.3 (Allerna Therapeutics Ltd). |
References
|