Research Article - Journal of Systems Biology & Proteome Research (2017) Journal of Systems Biology & Proteome Research (Special Issue-2017)
Human lysophosphatidic acid receptor 2-K1.31 and K7.36 gatekeeper functions provide new insight into robust affinity for LPA-type agonists.
Olaposi I Omotuyi*
Center for Bio-Computing and Drug Development, Adekunle Ajasin University, Akungba-Akoko, Ondo State Nigeria
- *Corresponding Author:
- Olaposi I Omotuyi
Center for Bio-Computing and Drug Development
Adekunle Ajasin University Nigeria
Tel: +234-80-709-43256
E-mail: olaposi.omotuyi@aaua.edu.ng
Accepted date: July 24, 2017
Citation: Omotuyi OI. Human lysophosphatidic acid receptor 2-K1.31 and K7.36 gatekeeper functions provide new insight into robust affinity for LPA-type agonists. J Syst Biol Proteome Res. 2017;1(1):1-4
Keywords
Lysophosphatidic acid receptor 2, Gatekeeper residues, Site-directed mutation, Calcium mobilization, Hydrophobic zipper, Robust affinity
Introduction
Since the discovery of lysophosphatidic acid (LPA) species as intercellular signal molecules rather than metabolic intermediates/products, (LPA) species have enjoyed an increasing research interest. As at today, at least eight (LPAR1- 5, P2Y5/10, GPR87) G-protein coupled receptor (GPCR) class [1] have been characterized as macromolecular entities conferring even increasing list of biological activities on LPA species. These biological functions border on cell survival and proliferation decisions [2] making them key players in many of clinical pathologies [3-5]. As mounting research evidence identify LPA/LPAR signaling in pathologies, chemical modulators of LPA receptors are now been viewed as key compounds in future clinical medicine.
LPA2, the second member of the family is an emerging key player in the treatment of hematopoietic and gastrointestinal acute radiation syndrome in mouse model of total-body irradiation [6] and a growing number of cancers [7]. Developing LPA2-specific drugs would be difficult as most LPA receptors share evolutionarily conserved orthosteric site residues; therefore, the robustness of the structural information we have on the receptors and the binding poses of their ligands would prove decisive if success is intended.
Two major reports have provided unique insights into the 3D structure of LPA1 in recent times; first, the crystal structure of LPA1 resolved in antagonist bound states [8] and the second report explored the roles of N-terminal K39 and intra-helical water path in ligand interaction [9]. Classical class-A GPCR inactivation pattern with the formation of transmembrane (TM)-III/VI ionic lock at the cytoplasmic end [10], closely packed N-terminal capping the 7 TM-helical bundle as seen in sphingosine -1-phosphate receptor 1 (S1PR1) and crystal waters in ligand interaction [11] were the consensus amongst the authors.
Taking key lessons from LPA1 structure as currently understood this study sought to understand why LPA2 has a higher affinity/ potency for the same LPA species in comparison with other LPA receptor subtypes [12] despite sharing evolutionarily conserved orthosteric residues (R3.28, Q3.29) [13]. To answer this question, homology modeling method and site-directed mutagenesis experiments were performed.
Materials and Methods
Homology modeling: Human lysophosphatidic acid receptor-2 (LPA2) sequence (REFSEQ: accession NM_004720.5) was retrieved from PubMed repository. LPA2 3D-model was built using LPA1 structure [8] on the SWISS-MODEL server after sequence alignment using DeepView (Swiss-PdbViewer).
Site-directed mutagenesis; the single or double mutants (K31A, R107A/Q108A, and E277A/K278A) were constructed using a QuikChange™ kit (Stratagene) with a plasmid containing an open reading frame that encodes the full-length LPA2 protein as the template for mutagenesis. The PCR reaction used Pfu DNA polymerase with the following primer pairs: K31A, 5′–CCA CTGGCGGCCCGCGGATGTGGTCGTGG–3′ (Forward) and 5′– CCACGACCACAT CCGCGGGCCGCCAGTGG–3′ (Reverse); R107A/Q108A, 5′– GGGCTGGTTCCTGGCGGCAGGCTTGCTGGACAC–3′ (F) and 5′–GTGTCCAGC AAGCCTGCCGCCAGGAACCAGCCC–3′(R); E 2 7 7 A / K278A, 5′– GTCCTGGCTGTAGCAGCGTACTTCCTACTG–3′ (F) and 5′–CAGTAGGAAGTA CGCTGCTACAGCCAGGAC–3′ (R). Each cycle involved heating the sample at 95 °C for 30 s, 55 °C for 1 min, and 68 °C for 2 min/kb of plasmid length; this sequence was repeated for a total of 16 cycles. The templates were digested with DpnI and transformed into E. coli HST08 premium competent cells (Takara). All mutations were confirmed by automated sequencing in both directions.
Cell culture and intracellular calcium ion (Ca2+) mobilization assay: B103 rat neuroblastoma cells that lack LPA response were cultured in DMEM containing 10 % fetal bovine serum at 370 C in a 5 % CO2 atmosphere. B103 cells were transfected with plasmid encoding either wild type or each mutant receptor using Lipofectamine. After 24 h of transfection, the cells were harvested by centrifugation and suspended with DMEM containing 10 % fetal bovine serum. The cell suspension was plated in a 384-well plate with the density of 1 × 104 cells/ well. Supernatant is removed and cells were incubated with serum-free media (DMEM/0.1% BSA) for 4 hours. Following incubation, cells were loaded with 10 μl Fluo-8 (8 μM: ABD Bioquest)) in 0.1% BSA supplied-DMEM containing 1 mg/ml amaranth. After 30 min, 20 μl of the LPA species at defined concentration was added followed by an immediate recording of the fluorescence using the Functional Drug Screening System/ μCell (Hamamatsu Photonics K.K., Hamamatsu city, Japan). The fluorescence intensity was described as Fluo-8 ratio (tested value/basal value) or fold induction. Western Blot assay for expressed/membrane localized LPA2 in wild-type and mutant conditions was done as previously described [9].
Results and Discussion
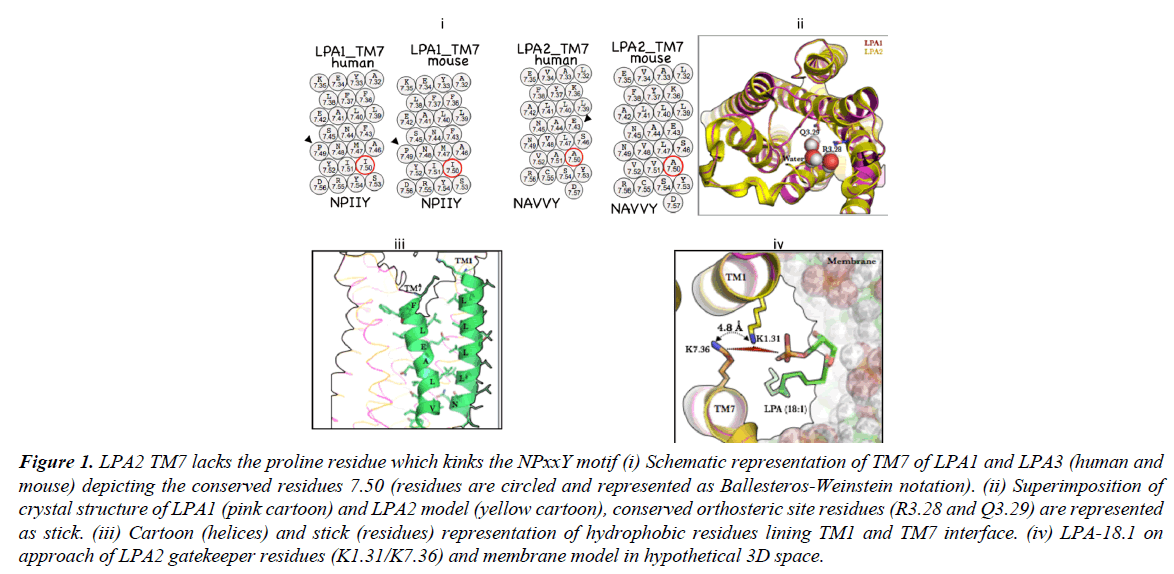
Agonists are more efficiently processed at LPA2 orthosteric site in comparison with other members of LPA receptors, which share high structural and evolutionary homologies i.e. LPA1, and LPA3. To understand this difference, focus was beamed on the mechanism of ligand approach to the receptor. It is now known that sphingosine-1-phosphate (S1P) and LPA species now approach their receptors by diffusing through a ligand channel located between TM1 and TM7 [13]. For classical GPCR-class A family, TM7 is known play a major role in ligand binding and receptor activation due to the presence of conserved K7.39 in electrostatic retention of highly charged LPA head group and conserved NPxxY motif which creates kink on the helix [8].
A notable difference between LPA1 and LPA2 primary sequence would be the absence of the P7.50 (NPXXY motif) in LPA2 and its replacement with A7.50 in human and mouse (Figure 1, i) Given that a kink at this position enabled by proline accounts for structural deformation required activation-deactivation dynamics [14] via a transmission switch mechanism [15] its absence in LPA2 will expectedly affects receptor activation. Furthermore, the role of TM7 in EDG-class GPCR activation is not restricted to function of P7.50 but may also play a more significant role in ligand recognition as speculated in the crystal structure of S1PR1 where the ligand is suspected to access the orthosteric tunnel by diffusing between TM1 and TM7. S1PR1-R7.34 has been suggested as the receptor gateway residue [16]
In order to understand how LPA2 deals with the absence of kink on the TM7, the 3D model was generated from LPA1 template (Figure 1, ii, superimposed structures of LPA1 (pink cartoon) and LPA2 (yellow cartoon). Interestingly the residues lining TM1 ((1.31)-KDVVVVALGLTVSVLVLLTNLL-(1.52)) and TM7 ((7.36)-KYFLLAEANSLVNAAVY-(7.53)) in LPA2 appear extensively hydrophobic (Figure 1, iii); whether they form hydrophobic zipper is not known. However, incontrovertibly, a hydrophobic zipper may restrict entry of LPA species into the orthosteric space thereby making receptor activation nearly impossible or when ligand gets into the orthosteric site, it makes escape nearly impossible thus, making activation more efficient.
Figure 1: LPA2 TM7 lacks the proline residue which kinks the NPxxY motif (i) Schematic representation of TM7 of LPA1 and LPA3 (human and mouse) depicting the conserved residues 7.50 (residues are circled and represented as Ballesteros-Weinstein notation). (ii) Superimposition of crystal structure of LPA1 (pink cartoon) and LPA2 model (yellow cartoon), conserved orthosteric site residues (R3.28 and Q3.29) are represented as stick. (iii) Cartoon (helices) and stick (residues) representation of hydrophobic residues lining TM1 and TM7 interface. (iv) LPA-18.1 on approach of LPA2 gatekeeper residues (K1.31/K7.36) and membrane model in hypothetical 3D space.
Our current understanding of GPCRs, which respond to lipid agonists, is that the N-terminal and the extracellular loops are closely packed with the transmembrane bundle thus covering the orthosteric site [16]. So how does LPA2 circumvent this challenge of hydrophobic zipper along the TM1/TM7 interface? In LPA2 an ingenious solution may have been evolutionarily provided; the presence of K1.31 whose location and orientation of its side chain from K7.36 allows for helical repulsion in the absence of an approaching ligand and cooperation during ligand approach, recognition and binding as the head group LPA species may bind stronger to two charged ε-amino groups of lysine (Figure 1, iv, showing LPA 18:1 approaching LPA1) as opposed to one lysine residue (K294) in LPA1.
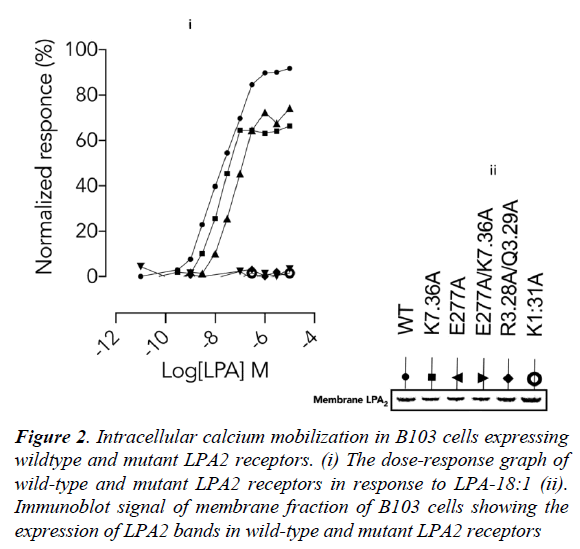
Next, the importance of these lysine residues in physiological condition was determined using site-directed mutagenesis experiments in LPA2 (wild-type and mutants) expressed in B103 cells. Wild-type LPA2 mobilized intracellular calcium ion in response to LPA-18:1 (Figure 2, i) which was completely abolished in orthosteric site (R3.28A/ Q3.29A) [13] double mutant. Interestingly, the response was also completely abolished in K1.31A (Figure 2, i) mutant. Furthermore, while K7.36A [13] and E277A mutation (Figure 2, i) only partially decreased the efficiency of LPA (18:1). A double mutant involving the residues (E277A/ K7.36A) completely abolished LPA-18:1 mediated LPA2 activation (Figure 2, i). These data therefore established that K1.31/E277 can partner howbeit inefficiently under K7.36A condition but neither K7.36 not E277 can function without K1.31.
Furthermore, the proximity of E277 to K7.36 strongly indicates that there may be a possible interaction between these residues as indicated by the mutagenesis data. A suggested role may be to assist K7.36 in the discharge of the ligand into the ligand tunnel while transiently replacing the ligand via a salt-bridge. It is worthy of note that the difference in calcium mobilization in the wild-type and mutant LPA2 is independent of gene transcription, translation and membrane localization as the membrane fractions from B103 cells showed similar band densities (Figure 2, ii).
Figure 2: Intracellular calcium mobilization in B103 cells expressing wildtype and mutant LPA2 receptors. (i) The dose-response graph of wild-type and mutant LPA2 receptors in response to LPA-18:1 (ii). Immunoblot signal of membrane fraction of B103 cells showing the expression of LPA2 bands in wild-type and mutant LPA2 receptors
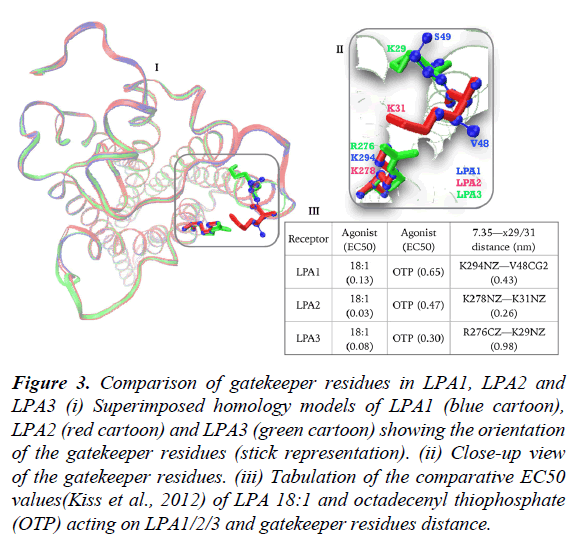
Lastly, we sought to study the contribution of gatekeeper residues to LPA3 as most of the discussion is centered on comparing LPA1 and LPA2 in terms of activation by LPA-type agonist. Noting that LPA3 also has NPxxY motif (data not shown), which conformationally regulates the opening, and closure of TM1/TM7 as present in LPA1. Superimposed homology models of LPA1 (blue), LPA2 (red) and LPA3 (green) built on LPA1 crystal structure (figure 3, i) revealed a fundamental orientation difference between LPA2 and LPA3 gatekeeper residues (figure 3, ii), first, R276/K29 pair in LPA3 is spaced at 0.98 nm apart (figure 3, iii) in contrast to 0.26 nm in LPA2 and K29 side chain in LPA3 assumes intra-helical orientation (figure 3, ii). To benefit maximally from the gatekeeper contribution, large conformational changes in TM1 helix aimed at relocating K29 side chain to TM1/TM7 interface may be required which ultimately explains the findings that LPA2>LPA3>>LPA1 in terms of response to LPA 18:1 (Figure 3, iii) [17].
Figure 3: Comparison of gatekeeper residues in LPA1, LPA2 and LPA3 (i) Superimposed homology models of LPA1 (blue cartoon), LPA2 (red cartoon) and LPA3 (green cartoon) showing the orientation of the gatekeeper residues (stick representation). (ii) Close-up view of the gatekeeper residues. (iii) Tabulation of the comparative EC50 values(Kiss et al., 2012) of LPA 18:1 and octadecenyl thiophosphate (OTP) acting on LPA1/2/3 and gatekeeper residues distance.
Conclusion
Robust LPA/LPA2 affinity can be explained by two major factors: 1) charge repulsion between K1.31 and K7.36, which keeps ligand tunnel open in perpetuity. 2) Tight binding between LPA-head group and the two ε-amino groups of the gatekeeping residues efficiently arrest ligand side and partitions it into the ligand tunnel. Additionally, E277 may provide needed mechanism for un-binding the ligand from K7.36 via salt-bridge formation while weakening the K1.31/ligand interaction
Conflicts of Interest
The author declares no conflict of interest.
Acknowledgements
This work was supported in part by grant to the Centre for Bio-Computing and Drug Development (CBDD) by Adekunle Ajasin University, Akungba-Akoko Bio-Medical Research Grant. First Bank Nigeria PLC also donated and installed the Hardware for Solar-Energy generation to CBDD under the CSR portfolio. Hardware for Computation was donated by Nagasaki University Advanced Computing Centre and Therapeutic Innovation Center of Nagasaki University Japan.
References
- Choi JW, Herr DR, Noguchi K, et al. LPA receptors: subtypes and biological actions. Annu. Rev. Pharmacol. Toxicol. 2010 Feb 10; 50:157-86.
- Ye X, Ishii I, Kingsbury MA, et al. Lysophosphatidic acid as a novel cell survival/apoptotic factor. BBA-MOL CELL BIOL L. 2002 Dec 30; 1585(2):108-13.
- Miyabe Y, Miyabe C, Iwai Y, et al. Necessity of lysophosphatidic acid receptor 1 for development of arthritis. Arthritis Rheumatol. 2013 Aug 1;65(8):2037-47.
- Ueda H, Matsunaga H, Olaposi OI, et al. Lysophosphatidic acid: chemical signature of neuropathic pain. BBA-MOL CELL BIOL L. 2013 Jan 31;1831(1):61-73.
- Yu X, Zhang Y, Chen H. Lysophosphatidic acid (LPA) stimulates invasion and metastatic colonization of ovarian cancer cells through Rac activation. Zhonghua Zhong Liu ZaZhi. 2015 Feb; 37(2):95-100.
- Deng W, Kimura Y, Gududuru V, et al. Mitigation of the hematopoietic and gastrointestinal acute radiation syndrome by octadecenylthiophosphate, a small molecule mimic of lysophosphatidic acid. Radiat. Res. 2015 Mar 25;183(4):465-75.
- Parrill AL. Design of anticancer lysophosphatidic acid agonists and antagonists. Future Med Chem. 2014 May; 6(8):871-83.
- Chrencik JE, Roth CB, Terakado M, et al. Crystal structure of antagonist bound human lysophosphatidic acid receptor 1. Cell. 2015 Jun 18;161(7):1633-43.
- Omotuyi OI, Nagai J, Ueda H. Lys39-lysophosphatidate carbonyl oxygen interaction locks LPA1 N-terminal cap to the orthosteric Site and partners Arg124 during receptor activation. Sci Rep. 2015; 5.
- Ballesteros JA, Jensen AD, Liapakis G, et al. Activation of the β2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J BIOL CHEM. 2001 Aug 3; 276(31):29171-7.
- Yuan S, Filipek S, Palczewski K, et al. Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat Commun. 2014 Sep 1; 5:4733.
- Hasegawa Y, Erickson JR, Goddard GJ, et al. Identification of a phosphothionate analogue of lysophosphatidic acid (LPA) as a selective agonist of the LPA3 receptor. J Biol Chem. 2003 Apr 4; 278(14):11962-9.
- Valentine WJ, Fells JI, Perygin DH, et al. Subtype-specific residues involved in ligand activation of the endothelial differentiation gene family lysophosphatidic acid receptors. J Biol Chem. 2008 May 2; 283(18):12175-87.
- Park JH, Scheerer P, Hofmann KP, et al. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008 Jul 10; 454(7201):183.
- Yuan S, Wu R, Latek D, et al. Lipid receptor S1P1 activation scheme concluded from microsecond all-atom molecular dynamics simulations. PLoSComput. Biol. 2013 Oct 3; 9(10):e1003261.
- Parrill AL, Lima S, Spiegel S. Structure of the first sphingosine 1-phosphate receptor. Sci Signal. 2012 May 22; 5(225):pe23.
- Kiss GN, Fells JI, Gupte R, et al. Virtual screening for LPA2-specific agonists identifies a nonlipid compound with antiapoptotic actions Mol. Pharmacol. 2012 Dec 1; 82(6):1162-73.