Research Article - Biomedical Research (2018) Computational Life Sciences and Smarter Technological Advancement: Edition: II
High performance angiogram sequence compression using 2D bi-orthogonal multi wavelet and hybrid speck-deflate algorithm
Somassoundaram T1*, Subramaniam NP2
1Department of Electronics and Communication Engineering, Sathyabama University, Chennai, Tamil Nadu, India
2Department of Electronics and Electrical Engineering, Pondicherry Engineering College, Tamil Nadu, India
- *Corresponding Author:
- Somassoundaram T
Department of Electronics and Communication Engineering
Sathyabama University, India
Accepted date: February 01, 2017
DOI: 10.4066/biomedicalresearch.29-16-2317
Visit for more related articles at Biomedical ResearchAbstract
Fast communication of medical information is a big challenge for today’s researchers. Either bandwidth or information needs to be sacrificed in order to communicate the entire information within a short time with minimal bandwidth. Compression of medical information before transmission helps us to communicate the entire information within short time with minimal cost. This paper concentrates on reducing the transmission bandwidth by compressing the entire medical information using a Hybrid algorithm which uses 2D Bi-orthogonal multi-wavelet transform and SPECK – Deflate encoder. The algorithm is studied with the standard test dataset GRUSELAMBIX. The real time data was analyzed to ensure the perseverance of the diagnostic information. 2D Bi-orthogonal multi-wavelet transform was applied on each frame on the DICOM file and the co-efficients are encoded using SPECK encoder followed by the deflate compression algorithm. To preserve the original diagnostic information, motion estimation and compensation was not applied on the medical videos. The qualitative performance of this algorithm is evaluated using Compression Ratio, PSNR, MSE, UIQIand SSIM. The Mean opinion Score from experts and non-experts was obtained on the real time data to evaluate the quantitative performance. The proposed algorithm revealed a better compression ratio than the traditional compression algorithm.
Keywords
Angiogram compression, Medical image compression, Multiwavelet compression, SPECK
Introduction
Digital Imaging and Communication (DICOM) and conservation of the medical diagnostic information have been an interesting research field for the recent years. Due to its larger file size and higher storage cost, the storage of the DICOM files is a big challenge. This also leads to the increase in the transmission cost. This paper mainly concentrates on reducing the storage and transmission cost for DICOM files by compressing as well as preserving the diagnostic information on the file. This paper proposes a hybrid three stage algorithm with multi-wavelet–SPECK and Deflate algorithm to achieve this goal.
This paper gives an overview of the proposed hybrid compression algorithm with three stages and also the quality assessment of the proposed algorithm is performed in the context of perseverance of medical diagnostic information. The quality assessment is studied in terms of Objective Quality and Subjective Quality. The subjective quality assessment is critical since the Subjective Quality needs to be considered while designing an algorithm to preserve the medical diagnostic information. Sometimes, the highly performed Algorithms in terms of objective quality may fail to perform in subjective quality assessment which leads to the loss of medical diagnostic information. Not all standards can be implemented on the files which contains the medical diagnostic information since the data manipulations carried out may distract the diagnosis. For an instance, the existing video coding standards that use the motion detection and estimation in the algorithm may disrupt the original information which leads to the false diagnosis [1]. Working with medical files requires an additional attention to preserve the diagnostic information.
In recent years, many compression algorithm techniques that use discrete wavelet transform has been proposed for medical diagnostic information such as MRI, CT Images, ECG signals, Ultra scan videos. In the previous work, a compression algorithm that uses Discrete Wavelet Transform and hybrid SPIHT deflate algorithm was proposed for medical images. Reduction in size of 50% was attained with less computation time [2]. Another compression algorithm that uses multiwavelet transform and hybrid SPIHT deflate algorithm was proposed for ECG signal which provided better compression ratio and PSNR [3]. Lossy compression technique for cardiac angiogram images was proposed by Minu et al. which uses the full frame DCT to preserve the fine details of the angiogram [4]. Ho et al. proposed a DWT based compression technique for angiogram files which uses motion compensation prediction to remove the inter frame correlations [5]. It is found that the use of DWT to the coronary angiogram files provides high quality and high compression. Gibson et al put forth a wavelet based compression technique for angiogram files in which the diagnostically significant areas are allocated with more number of bits. The coefficients are encoded using 3D- SPIHT [6,7].
Zaid et al. developed an algorithm with low computational cost based on H.264/AVC standard. In their algorithm the coding stage is enhanced by integrating a classification process that labels each fixed size region in the image as relevant or irrelevant and encode it accordingly [1]. Razaak et al. studied the quality assessment for medical videos compressed using HEVC using latest objective and subjective qualities [8]. The rest of the paper is as follows. In Section 2, the hybrid three stage algorithms with Multi wavelet – SPECK and Deflate algorithm along with overview of multiwavelet, SPECK algorithm is presented. In Section 3, a brief overview of the quality metrics taken for the assessment is provided. In Section 4, the simulation results and comparative results have been discussed and in Section 5, the paper is concluded.
Hybrid Three Stage Algorithm with Multiwavelet – SPECK and Deflate Algorithm
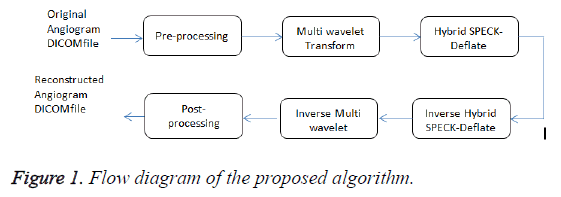
The hybrid algorithm with multiwavelet – SPECK and Deflate algorithm is better explained using Flow diagram (Figure 1). Unlike the normal images, the angiogram DICOM file contains numerous frames similar to video files. These DICOM file is pre-processed by splitting the file into frames and is then filtered to remove various noises caused while acquisition. The pre-processed filtered DICOM output is decomposed using multiwavelet transform. Then the coefficients are encoded using SPECK encoder. The encoded output is compressed using deflate algorithm. The compressed DICOM frames are again post-processed similar to original DICOM file by combining the frames. The storage size of the file is significantly reduced leading to minimum storage space. To reconstruct the original DICOM file, the compressed encoded DICOM file is decompressed using deflate algorithm and then the output is decoded with SPECK and inverse multiwavelet transform is applied. The reconstructed signal provides a better PSNR and preserves the diagnostic information.
An overview of multiwavelet transform and hybrid SPECK - deflate algorithm is provided below for the better understanding of the proposed algorithm.
Multi-wavelet transform
Multi-wavelet has been evolved from generalization of scalar vectors [9,10]. Multi wavelet transform uses multi scaling and multiple wavelet functions. Multi-wavelet shows an outstanding performance in terms of linear phase symmetry for preserving the boundaries, orthogonality of filters and vanishing moments for higher order of approximation [9]. The DWMT is well documented in the literature [9-11], so an overview is presented here.
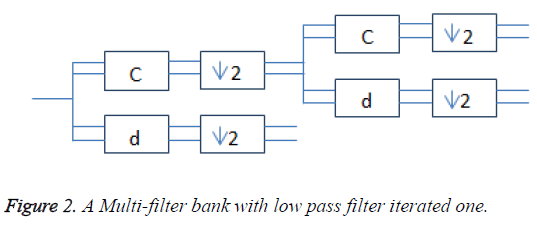
Multi-wavelets are mainly orthogonal and bi-orthogonal. The incoming scalar signal is converted into vector by using the pre-filter. Multiple high pass and multiple low pass filters were used to compute the discrete multiwavelet transform. The low pass filter (G) uses the low pass filter coefficients (Gk) and down sampled by 2 to get Hk coefficients. The multi-scaling function and the associated multi-wavelet function are given using (1) and (2). These two scale equations are realized using matrix filter bank as shown in Figure 2 operating on r input data streams and filtering them into 2r output data streams. Each is down sampled by the factor two.
Φ(t) = Σ Gk(2t-k) → (1)
W(t) = Σ Hk(2t-k) → (2)
Set partitioning embedded block coder (SPECK)
SPECK is the most popular algorithm used in image compression proposed by Pearlman et al. A brief overview of SPECK is provided here since SPECK is well documented in [12,13]. Unlike SPIHT, SPECK offers comparable results with lower complexity. Similar to SPIHT, there are three stages in SPECK- initialization, sorting and refinement. SPECK sorts the wavelet coefficients in terms of List of Significant Sets (LIS) and List of Significant Pixels (LSP).A start threshold (chosen as a power of two: T=2(n-1)) depending on the maximum value in the wavelet coefficient pyramid is defined at the initialization stage. The list LSP is set as empty, the image is then partitioned into two groups: set of type S which is the root of the pyramid and set of type Γ which is the rest of the image. A block of set of type S is determined by the coordinates of the pixel in the top-left and the size of this block. First, set of type S is added to the LIS. In the sorting pass, the algorithm first starts to sort each block of type S in LIS by performing a significant test against the current threshold either 1 or 0. A block is said significant if there is at least one coefficient in this block whose magnitude is greater than or equal to the threshold. If a block of type S is significant, it is partitioned into four subsets of the same type (S0, S1, S2 and S3).
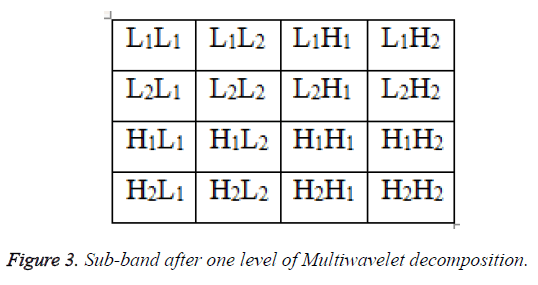
In the LIS, this block is replaced by the resulting subsets. In the case of a significant block of size 1 × 1 (one pixel), its sign is coded and then its co-ordinate is moved to the LSP. In the same way, the set Γ is tested with respect to the current threshold where its split up produces one subset Γ and three subsets of type S (S1, S2and S3) as depicted in Figure 3. This significance and partitioning process is carried out for all sets of type S (including the new ones) and the set Γ. At a certain point, the set Γ disappears depending upon the information content of the image and the desired bit-rate of coding. In the refinement pass, the nth most significant bit of each entry in the LSP, excluding those which have been added during the last sorting pass, is output. Then, the current threshold is divided by 2 and the sorting and refinement stages are continued. In the proposed algorithm, to obtain higher compression and to reduce the length to avoid the redundancy bits, the output of the SPECK encoder is processed with deflate algorithm which takes the advantage Huffman and LZW technique [2].
Quality Metrics
In this paper, objective quality and subjective quality have been assessed for the proposed hybrid algorithm with multiwavelet – SPECK and Deflate algorithm. Objective quality measure may not concentrate on preserving the diagnostic information of the medical data. To ensure the preservation of medical diagnostic information, the result for the subjective quality has been assessed through the opinion of a medical expert.
Objective quality metrics
Objective quality metrics can be broadly classified into statistical measure and human visual system measure. Each metrics is briefed below.
Statistical measure: The statistical measure is obtained based on the pixel information. Mean Square Error (MSE) is the cumulative squared error between the compressed image and the original image given using eq. (3). Peak Signal to Noise Ratio (PSNR) is the peak of the measured error given using eq. (4)
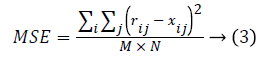
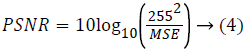
Where r refers to original image, x denotes restored image. The size of the image is M × N.
Human visual system measure: Structural Similarity Index Model(SSIM) is one of the metrics to measure the human visual system. It is developed to measure the difference between the original image and the distorted image in terms of structural information, luminance and contrast based on the human eye perception. SSIM is given using eq. (5)
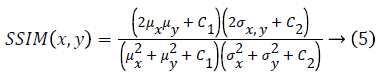
Where μx is the average of x obtained by
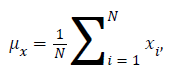
μy is the average of y obtained by
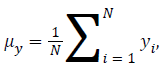
Standard Deviation of x obtained by
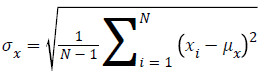
Standard Deviation of y given by
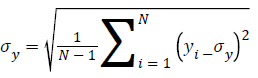
C1=(K1L)2, C2=(K2L)2 are two variables to stabilize the division with weak denominator; L the dynamic range of the pixel-values (for an 8 bit image it takes from 0 to 255), K1=0.01 and K2=0.03 by default.
Subjective quality metrics
The Subjective quality is finding the presence of medical diagnostic information on the output of the proposed algorithm. This assessment is obtained from the opinion of the expert on the quality of the image. The mean opinion score (MOS) has been used for subjective quality assessment. MOS is obtained by averaging the result of set of standard subjective tests performed for the proposed algorithm. MOS is a 5 point scale with the least as 1 which has no diagnostic information present in the output and best is 5 which contains the entire diagnostic information.
Simulation Results and Discussion
Some experiments have been performed to test the efficiency of the algorithm. The algorithm was developed in MATLAB 7.14.0.739. The performance of the hybrid three stage algorithms with multiwavelet – SPECK and Deflate algorithm is tested with GRUSELAMBIX -Standard DICOM image available in Osirix [14]. Bi-orthogonal wavelet families have been concentrated for this experiment.
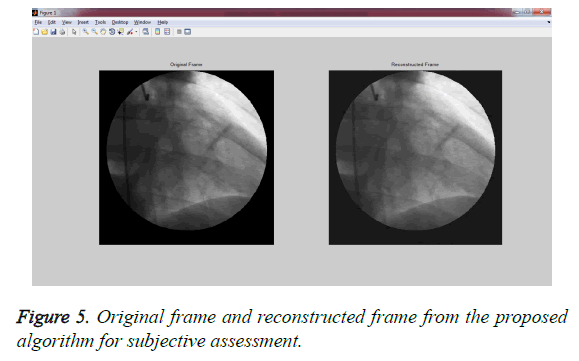
Experiment 1: This experiment was accomplished by performing the subjective evaluation of the hybrid algorithms with multiwavelet – SPECK and Deflate algorithm without applying any pre-processing filters on GRUSELAMBIX to evaluate the performance on clinical diagnosis information of the reconstructed signal. In this experiment, Mean Opinion Score (MOS) in five point scales was obtained from a group of medical experts and non- medical experts by evaluating the original signal and reconstructed signal through a questionnaire. Figure 4, shares the information obtained from the users (experts) through a questionnaire used for gathering the MOS. The assessment of Figure 5 shows the reconstructed image had preserved the medical diagnosis information in the original signal.
Subject evaluated: Video Sequence (Ultrasound, Angiogram)/ Standalone Image (T1, T2, X-ray).
Qualitative analysis
5- Excellent (Contains all information),
4- Very Good (Contains Diagnostic Information),
3- Good (Contains predictable Diagnostic information),
2- Bad (Little Diagnostic information),
1-Very Bad (No Diagnostic Information)
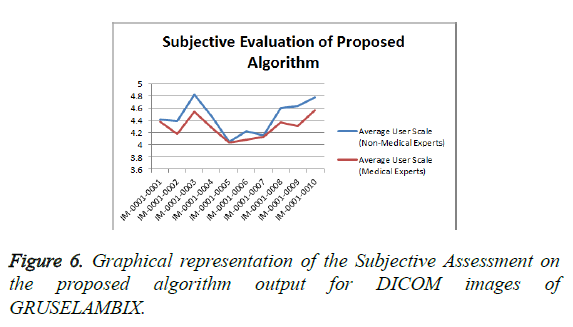
From the Table 1 it is seen that the Mean Opinion Score is above 4 which states that the Clinical Diagnostic Information is preserved. Figure 6 shows the Average User Scale of proposed algorithm for various DICOM images of GRUSELAMBIX.
| S. No | DICOM Image | Average User Scale (Non-Medical Experts) | Average User Scale (Medical Experts) |
|---|---|---|---|
| 1 | IM-0001-0001 | 4.41 | 4.38 |
| 2 | IM-0001-0002 | 4.39 | 4.18 |
| 3 | IM-0001-0003 | 4.82 | 4.54 |
| 4 | IM-0001-0004 | 4.47 | 4.29 |
| 5 | IM-0001-0005 | 4.05 | 4.04 |
| 6 | IM-0001-0006 | 4.23 | 4.08 |
| 7 | IM-0001-0007 | 4.15 | 4.13 |
| 8 | IM-0001-0008 | 4.6 | 4.37 |
| 9 | IM-0001-0009 | 4.63 | 4.31 |
| 10 | IM-0001-0010 | 4.78 | 4.57 |
Table 1: Subjective Assessment on the proposed algorithm for DICOM images of GRUSELAMBIX.
Experiment 2: This experiment was accomplished by performing the subjective evaluation of the hybrid algorithm with Multiwavelet – SPECK and Deflate algorithm on five real time angiogram DICOM files of the patients from Puducherry in India, in order to validate the performance on medical diagnosis information of the reconstructed signal. In this experiment, a five point Mean Opinion Score(MOS) was obtained from a group of medical experts and non- medical experts by evaluating the original signal and reconstructed signal through a questionnaire. Figure 4, shows the information obtained from the experts through a questionnaire used for gathering the MOS. The DICOM 3 belongs to a patient who is not normal and needs a medical assistance. Even from the reconstructed signal the same is diagnosed which clearly reveals that the clinical diagnostic information is preserved.
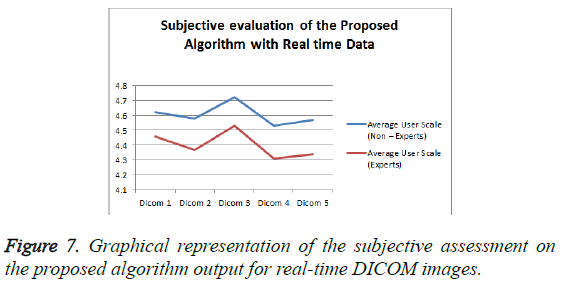
From the Table 2 it is seen that the Mean Opinion Score is above 4.5 which clearly states that the clinical Diagnostic Information is retained. Figure 7 represents the Average User Scale of the proposed algorithm for various real-time DICOM images.
| S. No | DICOM Image | Average User Scale (Non ? Experts) | Average User Scale (Experts) |
|---|---|---|---|
| 1 | DICOM 1 | 4.62 | 4.46 |
| 2 | DICOM 2 | 4.58 | 4.37 |
| 3 | DICOM 3 | 4.72 | 4.53 |
| 4 | DICOM 4 | 4.53 | 4.31 |
| 5 | DICOM 5 | 4.57 | 4.34 |
Table 2: Subjective Assessment on the proposed algorithm for real time Angiogram DICOM Files.
Experiment 3: The proposed algorithm is validated with various DICOM files of Coronary Angiogram GRUSELAMBIX in a study by Osirix [14]. Objective analysis is performed to verify the efficiency of the proposed algorithm.
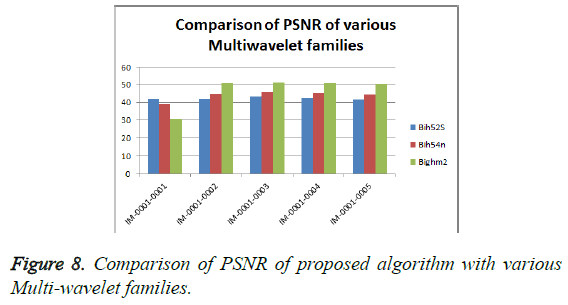
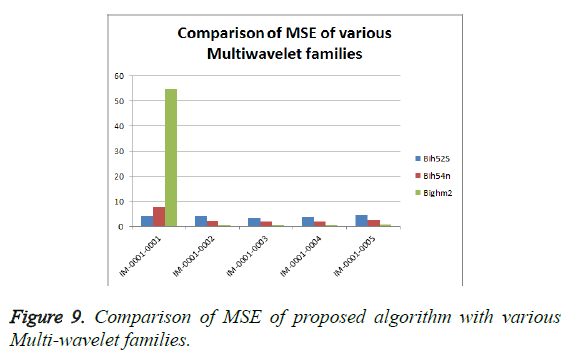
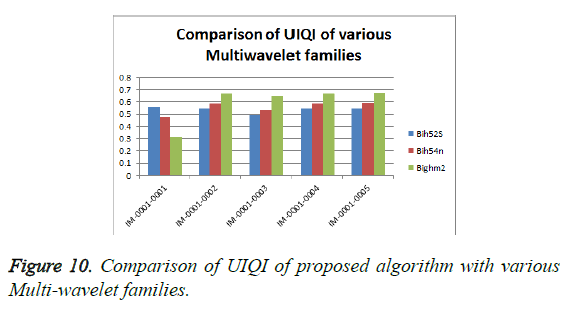
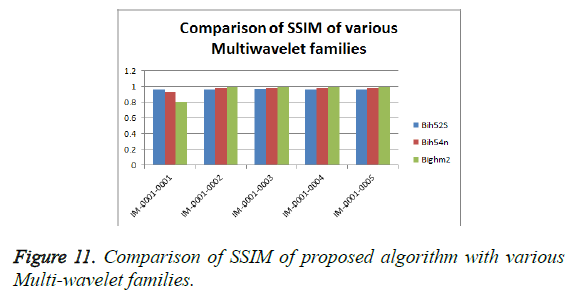
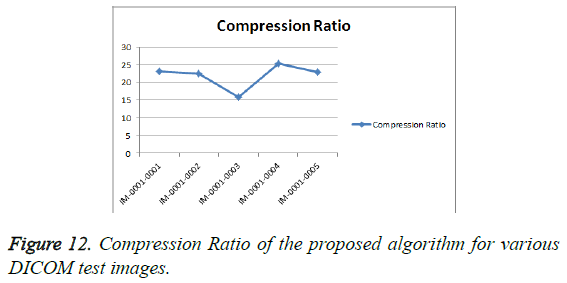
From Table 3, it is seen that the Bighm2 outperforms the others in the multiwavelet family by providing better performance evaluation. Figures 8-11 show the comparison of performance of proposed algorithm with various Multiwavelet families and it is seen that the Bighm2 provides better results in terms of PSNR, SSIM and Universal Image Quality Index (UIQI). Figure 12 shows the graphical representation to compression ratio of the proposed algorithm for the various DICOM images of GRUSELAMBIX.
| S. No | Wavelet | DICOM | PSNR | MSE | Universal Image Quality Index | SSIM | CR |
|---|---|---|---|---|---|---|---|
| 1 | Bih52S | IM-0001-0001 | 42.00 | 4.10 | 0.55891 | 0.95763 | 23.1494 |
| 2 | bih54n | IM-0001-0001 | 39.37 | 7.52 | 0.47898 | 0.92808 | 23.1494 |
| 3 | Bighm2 | IM-0001-0001 | 30.76 | 54.62 | 0.31324 | 0.80852 | 23.1494 |
| 4 | Bih52S | IM-0001-0002 | 41.89 | 4.21 | 0.54578 | 0.95676 | 22.5016 |
| 5 | bih54n | IM-0001-0002 | 44.69 | 2.21 | 0.58946 | 0.97331 | 22.5016 |
| 6 | Bighm2 | IM-0001-0002 | 50.79 | 0.54 | 0.67126 | 0.99262 | 22.5016 |
| 7 | Bih52S | IM-0001-0003 | 43.07 | 3.21 | 0.49685 | 0.96784 | 15.7501 |
| 8 | bih54n | IM-0001-0003 | 45.84 | 1.70 | 0.53558 | 0.97910 | 15.7501 |
| 9 | Bighm2 | IM-0001-0003 | 51.44 | 0.47 | 0.64445 | 0.99341 | 15.7501 |
| 10 | Bih52S | IM-0001-0004 | 42.52 | 3.64 | 0.54770 | 0.96236 | 25.2450 |
| 11 | bih54n | IM-0001-0004 | 45.25 | 1.94 | 0.58911 | 0.97637 | 25.2450 |
| 12 | Bighm2 | IM-0001-0004 | 51.00 | 0.52 | 0.66834 | 0.99286 | 25.2450 |
| 13 | Bih52S | IM-0001-0005 | 41.58 | 4.52 | 0.54934 | 0.95470 | 22.7694 |
| 14 | bih54n | IM-0001-0005 | 44.47 | 2.32 | 0.59529 | 0.97276 | 22.7694 |
| 15 | Bighm2 | IM-0001-0005 | 50.70 | 0.55 | 0.67495 | 0.99263 | 22.7694 |
Table 3: Objective Quality assessment of the proposed algorithm with various multiwavelet families applied to speck encoder for multiple DICOM images of GRUSELAMBIX.
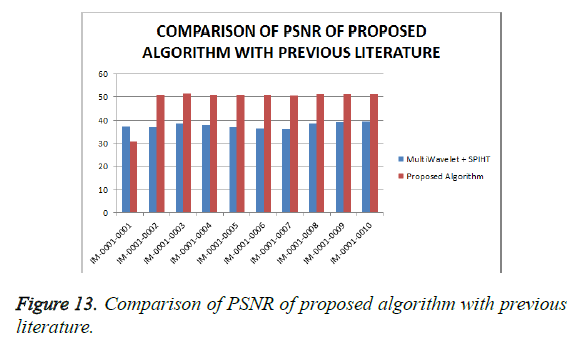
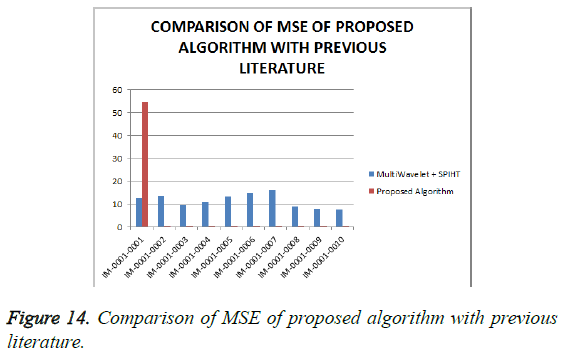
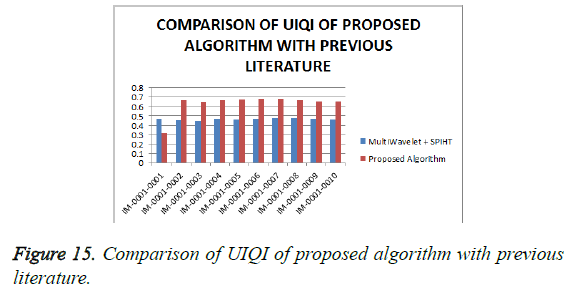
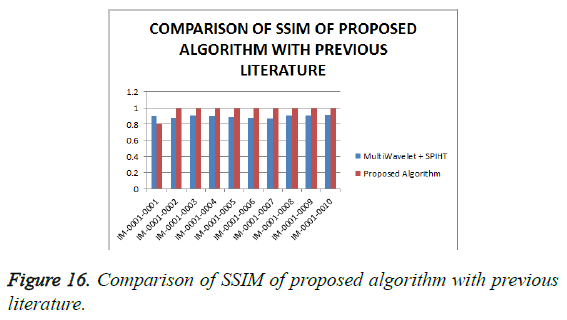
From Table 4, it is seen that the proposed algorithm outperforms the existing algorithm by providing better Objective Assessment. Figures 13-16 show the performance of the proposed algorithm yields a better performance compared to the existing algorithm.
| S. No | DICOM | Wavelet | PSNR | MSE | Universal Image Quality Index | SSIM |
|---|---|---|---|---|---|---|
| 1 | IM-0001-0001 | Multiwavelet+SPIHT | 37.13 | 12.58 | 0.46908 | 0.89284 |
| Proposed Algorithm | 30.76 | 54.62 | 0.31324 | 0.80852 | ||
| 2 | IM-0001-0002 | Multiwavelet+SPIHT | 36.79 | 13.61 | 0.45610 | 0.87754 |
| Proposed Algorithm | 50.79 | 0.54 | 0.67126 | 0.99262 | ||
| 3 | IM-0001-0003 | Multiwavelet+SPIHT | 38.41 | 9.39 | 0.44870 | 0.90676 |
| Proposed Algorithm | 51.44 | 0.47 | 0.64445 | 0.99341 | ||
| 4 | IM-0001-0004 | Multiwavelet+SPIHT | 37.80 | 10.79 | 0.47193 | 0.89598 |
| Proposed Algorithm | 51 | 0.52 | 0.66834 | 0.99286 | ||
| 5 | IM-0001-0005 | Multiwavelet+SPIHT | 36.90 | 13.27 | 0.45841 | 0.88665 |
| Proposed Algorithm | 50.7 | 0.55 | 0.67495 | 0.99263 | ||
| 6 | IM-0001-0006 | Multiwavelet+SPIHT | 36.38 | 14.97 | 0.46846 | 0.87582 |
| Proposed Algorithm | 50.70 | 0.55 | 0.67950 | 0.99288 | ||
| 7 | IM-0001-0007 | Multiwavelet+SPIHT | 36.05 | 16.16 | 0.47446 | 0.87255 |
| Proposed Algorithm | 50.64 | 0.56 | 0.68271 | 0.99307 | ||
| 8 | IM-0001-0008 | Multiwavelet+SPIHT | 38.57 | 9.05 | 0.48026 | 0.90381 |
| Proposed Algorithm | 51.05 | 0.51 | 0.66420 | 0.99332 | ||
| 9 | IM-0001-0009 | Multiwavelet+SPIHT | 39.18 | 7.86 | 0.46656 | 0.90601 |
| Proposed Algorithm | 51.10 | 0.51 | 0.65152 | 0.99298 | ||
| 10 | IM-0001-0010 | Multiwavelet+SPIHT | 39.28 | 7.67 | 0.45995 | 0.91148 |
| Proposed Algorithm | 51.14 | 0.50 | 0.65025 | 0.99309 |
Table 4: Comparison of performance evaluation for various DICOM images of gruselambix with Previous Literature.
Conclusion
In this paper, a simple and efficient compression technique for Coronary Angiogram using Multiwavelet transforms and Hybrid SPECK- Deflate has been discussed. The main goal of this paper is to reduce the transmission cost by reducing the storage space required for DICOM files through better compression ratio and better performance. Objective and Subjective assessment is performed on the reconstructed image to study the efficiency of the proposed algorithm. From the subjective analysis, the MOS is greater than 4 which exhibits the proposed algorithm is efficient to preserve the medical diagnostic information. Subjective assessment made with the real time data clearly exhibits that the diagnostic information is preserved in the algorithm. This research will be focusing further to reduce the computation time and from the analysis, the proposed algorithm with Bighm2, multi-wavelet type outperforms the other existing algorithms by providing superior compression ratio and superior performance in terms of PSNR, MSE, UIQI and SSIM through which the goal of the paper is reached.
References
- Zaid AO, Fradj BB. Coronary Angiogram video compression for remote browsing and archiving applications. Comput Med Imaging Graphics 2010; 34: 632-641.
- Somassoundaram T, Subramaniam NP. A Hybrid Scheme for Medical Image Compression Using SPIHTand DEFLATE Technique. Int J Eng Res Technol 2014.
- Somassoundaram T, Subramaniam NP. High Performance ECG compression using 2D Bi-Orthogonal Multiwavelet and Hybrid SPIHT- DEFLATEAlgorithm. Int J Appl Eng Res 2015.
- Minu RI, Nagarajan G, Suresh A, Jayanthila Devi A. Cognitive Computational Semantic for high resolution image interpretation using artificial neural network. Biomed Res 2016; 27: S306-S309.
- Ho B, Tsai M, Wei J, Ma M, Saipetch P. Video compression of coronary angiograms based on discrete wavelet transform with block Classification. IEEE Transact Med Imaging 1996; 15: 814-823.
- Gibson D, Spann M, Woolley SI. Diagnostically Lossless 3D Wavelet Compression for Digital angiogram Video. Proceedings of Data Compression Conference, 2002.
- Gibson D, Spann M, Woolley S. A Wavelet-Based Region of Interest Encoder for the Compression of Angiogram Video Sequences. IEEE Transact Informa Technol Biomed 2004; 8: 103-113.
- Razaak M, Martini MG. A study on Quality Assessment for Medical Ultrasound video compressed via HEVC. IEEE J Biomed Health Informa 2014; 18: 1552-1559.
- Strang G, Strela V. Short Wavelets and Matrix Di-lation Equations. IEEE Transact Signal Process 1995; 43: 108-115.
- Strang G, Strela V. Orthogonal Multiwavelets with Vanishing Moments. J Optical Eng 1994; 33: 2104-2107.
- https://arxiv.org/ftp/arxiv/papers/1010/1010.4816.pdf
- Said A, Pearlman WA. A New Fast and Efficient Image Codec Based on Set Partitioning in Hierarchical Trees. IEEE Transact Circuits Syst Video Technol 1996; 6: 243-250.
- http://ieeexplore.ieee.org/abstract/document/1347192/
- http://www.osirix-viewer.com/datasets/