Research Article - Journal of RNA and Genomics (2016) Volume 12, Issue 1
Exploring Riboswitches in Archaeal Metagenomes
Angela Gupta1, and D Swati2*1Department of Bioinformatics, Mahila Mahavidyalaya, Banaras Hindu University, Varanasi, Uttar Pradesh, India
2Department of Physics, Mahila Mahavidyalaya, Banaras Hindu University, Varanasi, Uttar Pradesh, India
Received Date: 24 August 2016; Revised Date: 03 January 2017 ; Accepted Date: 11 January 2017; Published Date: 16 January 2017
Copyright: First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Abstract
Metagenomics is defined as the direct analysis of genomes found in an environmental sample. It involves cloning and analyzing the genomes without culturing the organisms in the community. A number of new and novel molecules with significant functionalities and applications have been identified through this approach. This study focuses on identifying riboswitches in Archaeal metagenomes. Instances of TPP and Ado-Cbl variant riboswitches have been identified in different uncultured archaeal metagenomes. These riboswitches were searched and verified by using various bioinformatics approaches. The findings in Archaeal metagenomes hint at the possibility of finding more predefined and novel classes of riboswitches as new genomic samples are extracted from the environment.
Keywords
Metagenome, comparative metagenomics, riboswitch, archaea, metabolism
Introduction
All the ecosystems, such as groundwater, soil, as well as the skin surface and the gastrointestinal tract of animals contain microbial communities that interact with each other and take part in the biological processes surrounding them. The habitat of a community has effects on the complex system of the microbial communities. Metagenomics is the study of genetic material recovered directly from such microbial communities residing in environmental samples. Metagenomics sheds light on the functioning of microbial communities and their surrounding ecosystem. In metagenomic studies, the genomic sequences of a collection of microorganisms are directly extracted from a specific environment. Among the methods designed to gain access to the physiology and genetics of uncultured organisms, metagenomics, the genomic analysis of a population of microorganisms, has emerged as a powerful method (Handelsman, 2004). For analyzing a metagenomic sequence data set, identifying functional sequence elements is a crucial step. Metagenomics is one of the fastest growing scientific disciplines. Originally conceived as a study of collective genomes of non-culturable microorganisms, metagenomics now refers to the use of high throughput DNA sequencing to provide taxonomic and functional profiles of microbial communities without the need to culture microbes in the laboratory (Franzosa et al, 2015).
Metagenomics provides access to the functional gene composition of microbial communities and thus gives a much broader description than phylogenetic surveys, which are often based only on the diversity of one gene, for instance the 16S rRNA gene. On its own, metagenomics gives genetic information on potentially novel biocatalysts or enzymes, genomic linkages between function and phylogeny for uncultured organisms, and evolutionary profiles of community function and structure (Thomas et al, 2012). Metagenomics is also a powerful tool for generating novel hypotheses of microbial function- the remarkable discoveries of proteorhodopsin-based photoheterotrophy or ammonia-oxidation in Archaea attest to this fact (Béja et al, 2000; Nicol and Schleper, 2006). By integrating the information so gleaned with the information about biological functions within the community, the habitat and functional aspects of microbial communities can potentially be probed. Metagenomics could also unlock the massive uncultured microbial diversity present in the environment to provide new molecules for therapeutic and biotechnological applications [http://www.nature.com/nrmicro/focus/metagenomics/index. html]. Recent discoveries in the area of genomics have revealed number of microRNAs (miRNAs) and riboswitches. The numbers of small RNAs (sRNAs) and small nucleolar RNAs (snoRNAs) have also been greatly expanded by recent analyses. However, discovering and analyzing functional RNA sequences is relatively more difficult as compared to more well-developed methodologies for discovering and analyzing protein coding sequences. This suggests the probability of some other RNAs that are likely to be undiscovered or unidentified.
Microorganisms residing in microbial communities help in survival of other microbes through symbiotic interaction. This type of association has been studied in Cenarchaeum symbiosum, a symbiont of a marine sponge, Axinella sp. (Preston et al, 1996). Comparative analyses between metagenomes can provide additional insight into the function of complex microbial communities, their interaction among each other and their role in their host health. Pairwise or multiple comparisons between metagenomes can be made at the level of sequence composition (comparing GC-content or genome size), taxonomic diversity or functional aspect.
This study is based on identifying riboswitches in the Archaeal archaea. Riboswitches are complex folded RNA elements that act as receptors for specific metabolites. They are found in the non-coding portions of various mRNAs, where they control gene expression via structural changes that are brought about by metabolite binding. Riboswitches play an important role in the regulation of bacterial transcription and translation without any involvement of proteins (Mandal and Breaker, 2004; Winkler and Breaker, 2005; Breaker, 2012). Riboswitches regulate several metabolic pathways, for example, the biosynthesis of vitamins (e.g. riboflavin, thiamin and cobalamin) and the metabolism of methionine, lysine and purines. Candidate riboswitches have also been observed in archaea and eukaryotes. Riboswitches are thought to be the descendants of an ancient sensory and regulatory system that likely functioned before the emergence of enzymes and genetic factors made of proteins (Strobel and Cochrane, 2007).
The study reveals the sporadic occurrence of riboswitches in Archaeal metagenomes, so far. However, their presence in other domains has been widely studied. The known riboswitches are involved in regulation of numerous fundamental metabolic pathways (Mandal et al, 2003; Vitreschak et al, 2004) and have been found not only in bacterial genomes, but also in archaeal and eukaryotic genomes (Sudarsan et al, 2003). Several metagenomic samples have been searched in this study from varied sources like hydrothermal vent sediments to hypersaline Antarctic lake. However, only two riboswitches have been identified in samples, TPP and Ado-Cbl Riboswitch. We have identified Thiamine pyrophosphate (TPP) and (Adenosylcobalamine) Ado-Cbl Variant riboswitches in Archaeal Metagenomes by using bioinformatics approaches. TPP represents one of the most intensively studied riboswitch class and is the most essential cofactor in bacteria, archaea, and eukatyotes. TPP is the active form of thiamine (vitamin B1), an essential coenzyme synthesised by coupling of pyrimidine and thiazole moieties in bacteria. TPP riboswitches were first identified in B. subtilis and E. coli.
AdoCbl riboswitch is a cis-regulatory riboswitch element present in untranslated regions of mRNA where binding adenosylcobalamin (Ado-Cbl) causes a change in gene expression. Ado-Cbl is a source of carbon-based free radicals that enzymes use to catalyze a variety of isomerizations (Rodionov et al, 2002). Ado-Cbl is one of two coenzymes derived from vitamin B12, the other being methylcobalamin (MeCbl) (Marsh 1998). Both coenzymes derive biological function from the ability to form a unique, stable, covalent bond between the central cobalt atom and 5’ carbon of adenosine (Fox et al, 2009). This riboswitch has been shown to be conserved across several strains of bacteria and has an influence over the expression of the ethanolamine utilization genes. It is proposed to be a subclass of the previously described classical Ado-Cbl-sensing riboswitch. This new subclass of Ado-Cbl riboswitches has both sequence and structural conservation with the core region of the classical Ado-Cbl-sensing riboswitches however it has some clear differences. The new element lacks a paired region formed by the pairing of the 5’-3’ ends found in canonical Ado-Cbl riboswitches. It also has an additional extra base-paired region near the 3′ terminus that is absent from the classical Ado-Cbl riboswitches (Banerjee, 2006).
Material And Methods
Genomic sequences of Archaeal metagenomes were obtained from NCBI Genome (http://www.ncbi.nlm.nih.gov/genome/). We also looked for specifically three genomes, Lokiarchaeum sp.GC14_75, isolated from Loki’s castle hydrothermal vent sediment, Gakkel Ridge, halophilic archaeon DL31, isolated from Deep Lake, an extremely cold and hypersaline Antarctic lake, and Candidatus Thorarchaeota archaeon SMTZ-45, isolated from Sulfate-methane transisition zone estuary sediments 16-26 cm, USA(White Oak River Estuary, North Carolina).
These metagenomic sequences were analysed for different riboswitches using bioinformatics approaches like BLAST Search, INFERNAL (http://eddylab.org/infernal/) and using descriptors in RNABOB program (ftp://ftp.genetics.wustl. edu/pub/eddy/software/). Riboswitch Scanner (http://service. iiserkol.ac.in/~riboscan/) (Mukherjee and Sengupta 2015) was used to analyze whole genomes for different riboswitches. Riboswitch Scanner is able to detect 24 classes of riboswitches from complete genome sequences as well as from partial genome sequences.We obtained hits for TPP riboswitch and Ado-cbl variant riboswitch. Hits of TPP riboswitch were further verified by using Ribopythia server (http://bioinformatics.bc.edu/ clotelab/RiboPythia/index.spy) [19]. Similarly, homologous sequences for Ado-cbl variant riboswitch were further analyzed by using BLAST search (https://blast.ncbi.nlm.nih.gov/Blast. cgi?PAGE_TYPE=BlastSearch).TPP riboswitch sequences from environmental samples were also used as seeds in BLAST search for finding TPP riboswitch in metagenomes (Kazanov et al, 2007). Their gene context was predicted by using NCBI. Identified hits of riboswitches were modelled using ModeRNA server (http://genesilico.pl/moderna/) (Rother et al, 2011). The 3-D structures were then compared with the known riboswitch structure using Pymol and analyzed.
Results
TPP riboswitch
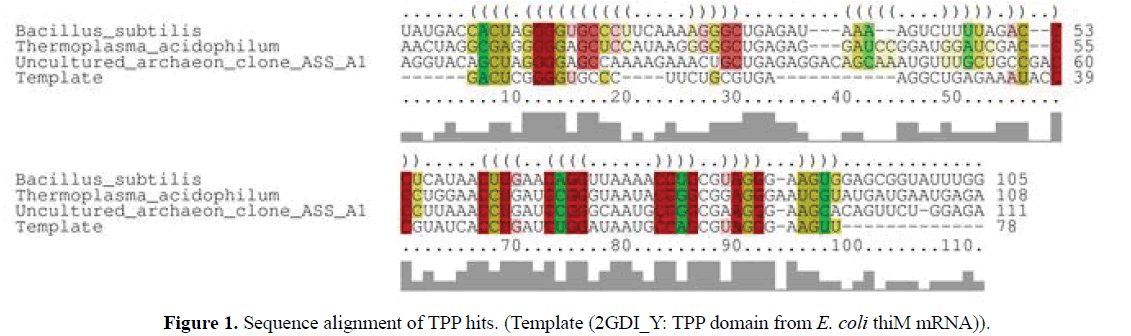
Instance of TPP riboswitch is found in uncultured archaeon clone ASS A1 isolated from acid sulfate soils of farmland in Australia, Murry Bridge from a depth of 0.80 metre. The hit was compared to an already defined riboswitch in Archaea, Picrophilus torridus. This organism is a thermoacidophile, and can grow at temperatures around 60oC, in environments where the pH drops to 0. The name reflects the area where the organism was isolated, which was a hot dry soil in Kawayu, in northern Japan, that was solfataric (volcanic area that gives off sulfur) (Fütterer et al, 2004) and was also compared to the bacteria,Bacillus subtilis subtilis. Based on sequence conservation, structure conservation and free energy, we concluded it as putative TPP riboswitch. TPP domain from E. coli thiM mRNA was used as template for the modeling of the TPP riboswitch from the uncultured archaeon clone ASS A1 (Table 1).
| S.no. | Organism | Freeenergy(dG=Kcal/mol) |
|---|---|---|
| 1 | Bacillus subtilissubtilis | -29.38 |
| 2 | Picrophilustorridus | -29.64 |
| 3 | Uncultured_archaeon_clone_ASS_A1 | -28.63 |
| 4 | Escherichia coli | -23.00 |
Table 1: Free energies of TPP riboswitches from different domains.
Comparative study of TPP riboswitch in archaea, archaeal metagenome and bacteria
TPP Riboswitch Sequences in different organism from different domains
>B_ subtilis _subtilis
U A U G A C C A C U - AGGGGUGCCCUUCAAAAGGGCUGAGAUAAAAGUCUUUUAGACCCUCAUAACUUGAACAGGUUAAAACCUGCGUA
GGGAAGUGGAGCGGUAUUUGG >Picrophilus_torridus
GUCGACUGGGGAGCCGAAAAGGCUGAGAGGAUCUAAAAAGAUCGACCCUUUGAACCUGAUCCGGGUAAUACCGGCGGAGGGAUU
UUAGUGGAAUUGAAU >Uncultured_archaeon_clone_ ASS_A1
A G G U A C A G C U A G G G G A G C - CAAAAGAAACUGCUGAGAGGACAGCAAAUGUUUGCUGCCGACCCUUAAACCUGAUCCGGGCAAUGC
CGGCGAAGGGAAGCACAGUUCUGGAGA
>Template (2GDI_Y: TPP domain from E. coli thiM mRNA)
GACUCGGGGUGCCCUUCUGCGUGAAGGCUGAGAAAUACCCGUAUCACCUGAUCUGGAUAAUGCCAGCGUAGGGAAGUU
Modelling of TPP riboswitch
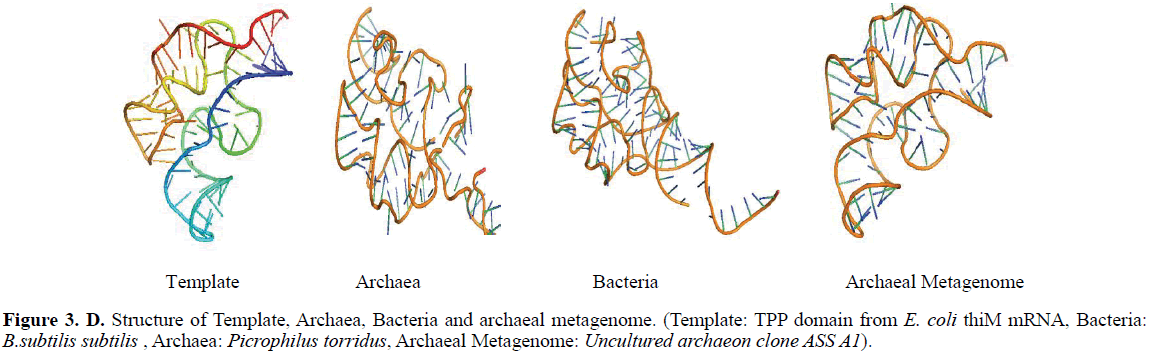
Using TPP riboswitch from E. coli as template, TPP sequence from archaea, bacteria and archaeal metagenome was modelled using ModeRNA server (Figure 1).
Comparative modeling of TPP riboswitch from archaea, bacteria and archaeal metagenome and template
Score was obtained by using ARTS web server (http:// bioinfo3d.cs.tau.ac.il/ARTS/pairwise_alignment.html) (Dror et al, 2006) (Table 2). It gives the information about (i) the number of matched base pairs (BP Core Size); (ii) the total number of matched nucleotides including unpaired ones (Core Size); (iii) the root mean square deviation (RMSD) between the phosphate atoms of the matched nucleotides in the core; (iv) the P-value.
| S.no. | Alignment | Score | BP coresize(nt) | Coresize(nt) | RMSD(Å) | P-value |
|---|---|---|---|---|---|---|
| 1 | Archaea and template | 96.00 | 12 | 72 | 0.91 | 0.0104 |
| 2 | Bacteria and template | 110.00 | 19 | 72 | 0.48 | 0.0068 |
| 3 | Metagenome andtemplate | 107.00 | 17 | 73 | 0.48 | 0.0078 |
| 4 | Picrophilustorridusand Metagenome | 95.00 | 13 | 69 | 1.06 | 0.0019 |
Table 2: Alignment score of TPP riboswitches from different sources.
Discussion
TPP riboswitch
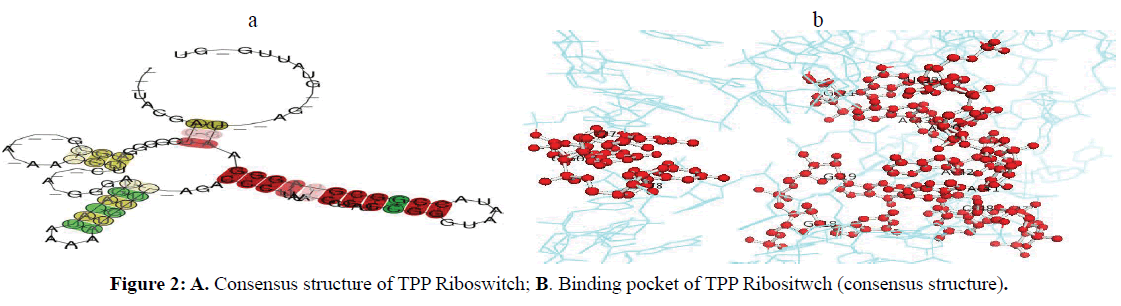
The binding arrangement of TPP in the binding pocket of TPP riboswitch is held by a T-loop-like turn formed by the conserved U39-G40-A41-G42-A43 segment, closed by a reverse Hoogsteen U39-A43 pair. The backbone is extended at both G40-A41 and G42-A43, resulting in the mutual intercalation and insertion of the Thiamine ring between these interacting segments (Figure 2A-B). Further, the fold is stabilized by continuous stacking of A41, G42, TPP, A43 and G21, and by the formation of (G19-A47)-G42 and (G18-C48)-A41 triples. The pyrophosphate group of TPP is bound in a spacious pocket by a pair of hexa-coordinated Mg2+ ions (Mg1 and Mg2) with octahedral ligation geometry that are positioned by direct and water mediated hydrogen bonds with RNA .The terminal phosphate of TPP is coordinated to both Mg1and Mg2, whereas the thiazole-linked phosphate is coordinated to Mg2. Mg1 is directly coordinated to the O6 carbonyls of the conserved G60 and G78 that are located in the region previously implicated in pyrophosphate recognition. In addition, the conserved C77 and G78 form hydrogen bonds to the oxygen atoms of the terminal phosphate of TPP (Serganov et al, 2006) (Figure 3).
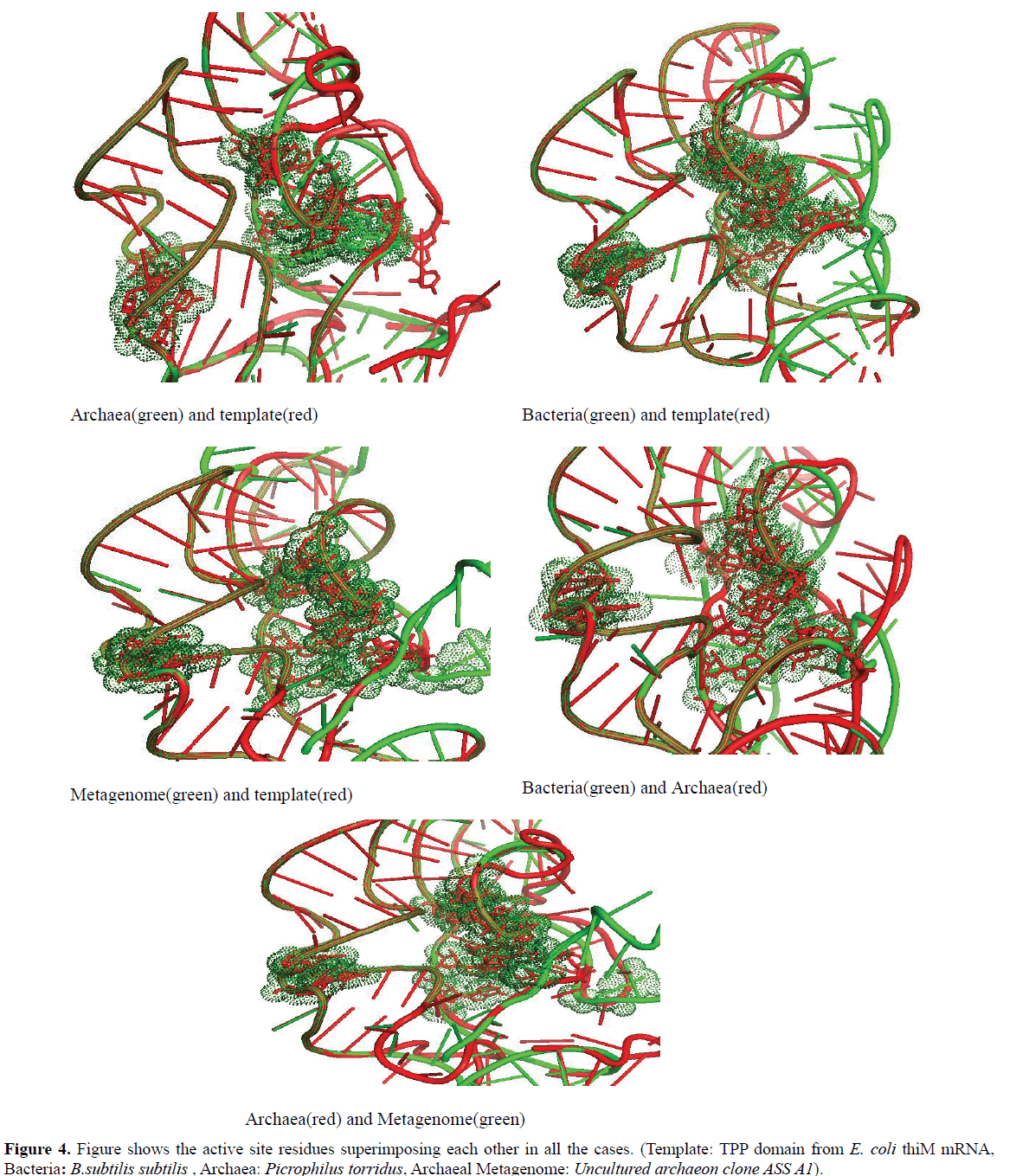
In this study it is observed that in every domain, the active site residues that respond to the TPP are well conserved (Figure 4).
We found that the active site residues of bacteria, archaea and archaeal metagenome TPP riboswitch were lying in the same proximity and were exactly superimposed on each other. This clearly indicates that TPP riboswitch domain shows significant level of conservation of the binding pocket among all the domains of life.
Ado-Cbl variant riboswitch
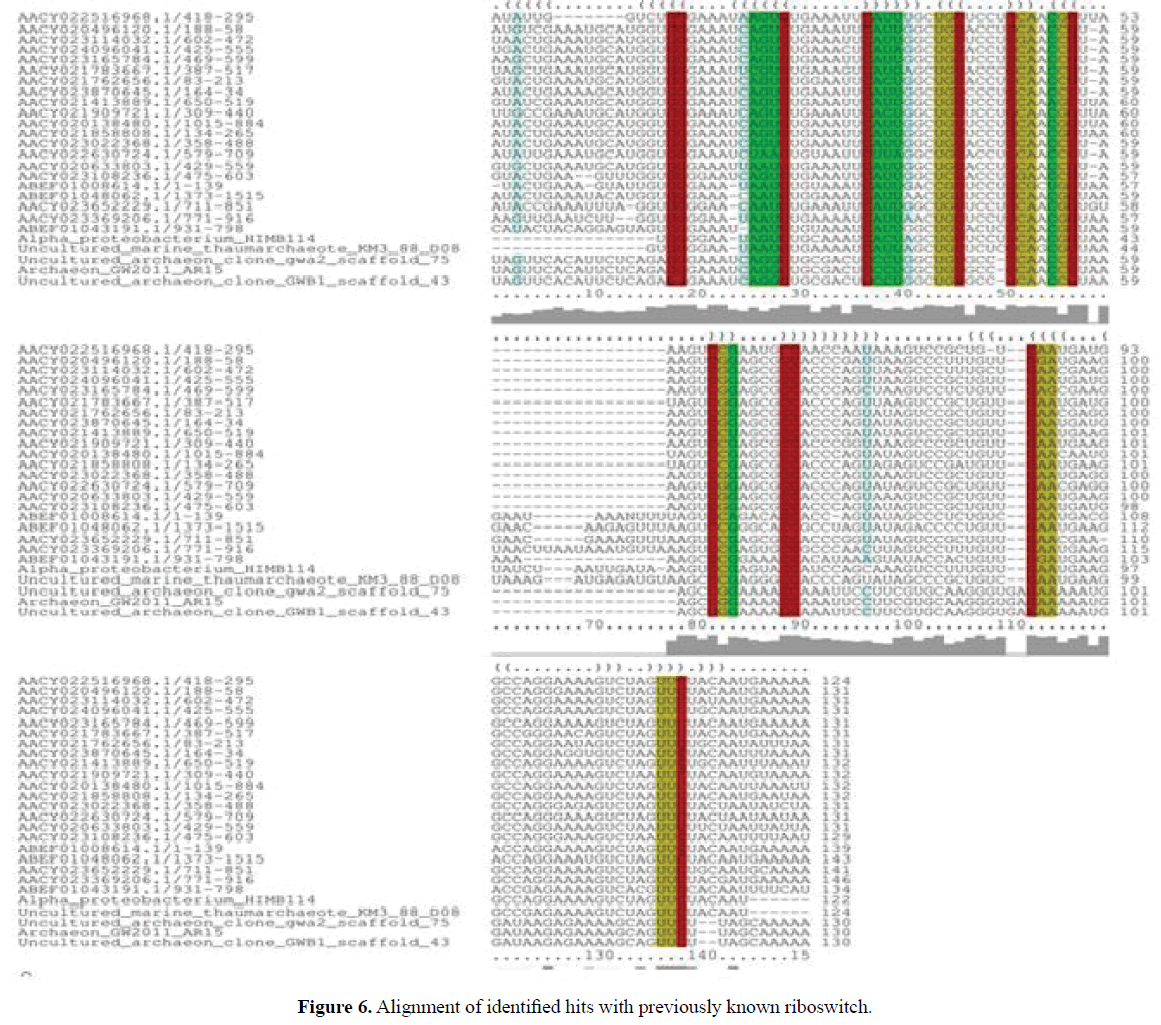
Hits for Ado-Cbl variant riboswitch were obtained in different environmental samples. Uncultured archaeon clone gwa2_ scaffold_75 genomic sequence and Uncultured archaeon clone GWB1_scaff old_43 genomic sequences were obtained from aquifer sediment, derived from Rifle groundwater metagenome (Table 3).
| S.no. | Organism | Freeenergy(dG=Kcal/mol) |
|---|---|---|
| 1 | Uncultured archaeon clone gwa2_scaffold_75 | -28.20 |
| 2 | Uncultured archaeon clone GWB1_scaffold_43 | -28.20 |
| 3 | Archaeon GW2011_AR15 | -28.20 |
| 4 | Uncultured marine thaumarchaeote KM3_88_D08 | -27.20 |
| 5 | Alpha proteobacterium HIMB114 | -26.60 |
Table 3: Free energy of Ado-Cbl variant riboswitch sequences.
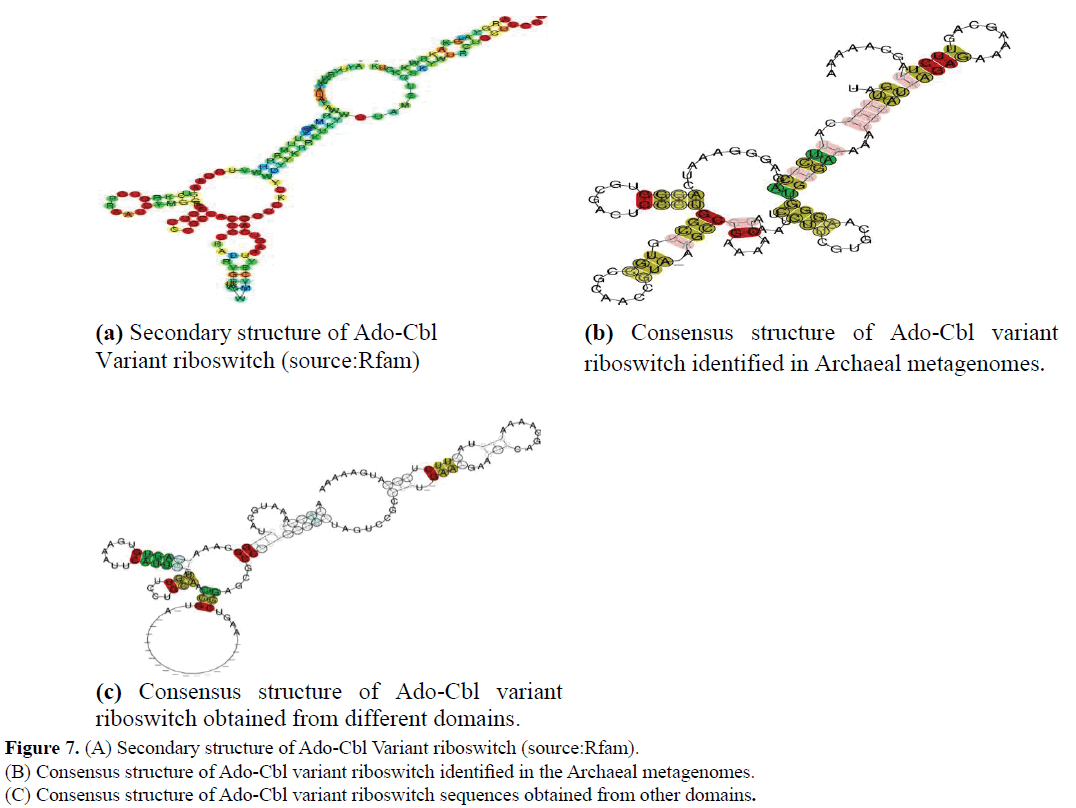
Uncultured marine thaumarchaeote KM3_88_D08 genomic sequence was obtained from Ionian sea and Archaeon GW2011_ AR15 genome was obtained from groundwater. Sequences were downloaded from NCBI genome. The sequences thus obtained were further analysed at their sequence and structural level by comparing them to previously defined riboswitch. Their sequence alignment among them and among other organisms derived from marine metagenomes (Source: Rfam) is given in the (Figures 5 and 6). Their secondary structures were compared with previously identified Ado-Cbl Variant riboswitch. It was observed that the secondary structures were very similar to the previously identified Ado-Cbl variant riboswitch and have a high degree of conservation among all the domains (Figure 7A-C).
Conclusion
Previous studies (Yadav et al, 2015) have established the TPP riboswitch to be widely distributed in bacteria, and quite abundantly in plants and fungi. However the samples of Archaeal metagenomes yielded only a few instances of TPP as revealed in this study, thereby pointing to the lesser utility of vitamin B metabolic factor in their metabolism. This is also corroborated by the search of TPP riboswitch in Archaeal genomes (manuscript under preparation).
These regulatory RNA elements participate actively in the gene regulatory processes. Also, in this study, other than Picrophilus torridus no more instances of TPP riboswitch were found in genomes obtained from sulphur rich environment. Other previously known instances of TPP riboswitches are found in different environmental conditions like Thermoplasma acidophilum, a thermophilic, acidophilic archaeon that was isolated from a self-heating coal refuse pile in south-western Indiana (Ruepp et al, 2000), Thermoplasma volcanium, isolated from acidic hydrothermal vents on the shore of Aeolian Island of Vulcano, Italy (Kawashima et al, 2000), Ferroplasma acidarmanus, an archeon growing at low pH in metal-rich solutions and is an iron-oxidizing organism capable of growth at pH 0 (Allen et al, 2007) and Candidatus Korarchaeum cryptofilum OPF8 growing in high temperatures (Elkins et al, 2008). This suggests a very sparse presence of occurrence of TPP riboswitch in sulphur rich condition.
Other than TPP, instances of Ado-Cbl variant riboswitch have also been identified in the environmental samples chosen. This suggests the possibility of identification of many more novel and variant form of riboswitches in metagenomes. We also studied three other unclassified archaeal genomes, Lokiarchaeum sp. GC14_75, halophilic archaeon DL31, Candidatus Thorarchaeota archaeon SMTZ-45, using three approaches, NCBI BLAST search, INFERNAL search, and RNABOB. We obtained few sequence hits of some riboswitches and ribozymes (Table 4). Although, their secondary structures did not match the secondary structure of established riboswitches, but getting similar sequences suggest a possibility of occurrence of variant forms of known riboswitches. Moreover, on the basis of diversity Other than TPP, instances of Ado-Cbl variant riboswitch have also been identified in the environmental samples chosen. This suggests the possibility of identification of many more novel and variant form of riboswitches in metagenomes. We also studied three other unclassified archaeal genomes, Lokiarchaeum sp. GC14_75, halophilic archaeon DL31, Candidatus Thorarchaeota archaeon SMTZ-45, using three approaches, NCBI BLAST search, INFERNAL search, and RNABOB. We obtained few sequence hits of some riboswitches and ribozymes (Table 4). Although, their secondary structures did not match the secondary structure of established riboswitches, but getting similar sequences suggest a possibility of occurrence of variant forms of known riboswitches. Moreover, on the basis of diversity and in establishing how widespread these classes are among organisms from the three domains of life.
| S. no. | HalophilicArchaeon DL31 | Lokiarchaeumsp. GC14_75 | CandidatusThorarchaeota archaeon SMTZ-45 |
|---|---|---|---|
| 1 | HHR II, III | HSP 90 | TPP |
| 2 | ykKc | Cobalamin | ykKc |
| 3 | Intron III | SAM 1 | Intron II |
| 4 | yYbP | Four U | Four U |
| 5 | ydaO | Purine | ykoK |
| 6 | FMN | HHR III | glmS |
| 7 | ykKc | FMN | |
| 8 | TPP |
Table 4: Sequence hits of some riboswitches/ribozymes obtained by three approaches in three unclassified archaea.
References
- Allen, Tyson, Whitaker, Detter, et al. 2007. Genome dynamics in a natural archaeal population. Proceedings of the National Academy of Sciences 1046, 1883-1888.
- Banerjee 2006. B12 trafficking in mammals: a case for coenzyme escort service. ACS chemical biology 13, 149-159.
- Béja, Aravind, Koonin, Suzuki, Hadd, Nguyen, et al. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 2895486, 1902-1906.
- Dror, Nussinov and Wolfson 2006. The ARTS web server for aligning RNA tertiary structures. Nucleic acids research 34suppl 2, W412-W415.
- Elkins, Podar, Graham, Makarova, et al. 2008. A korarchaeal genome reveals insights into the evolution of the Archaea. Proceedings of the National Academy of Sciences 10523, 8102-8107.
- Fox, Ramesh, Stearns, Bourgogne, et al. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proceedings of the National Academy of Sciences 10611, 4435-4440.
- Franzosa, Hsu, Sirota-Madi, Shafquat, et al. 2015. Sequencing and beyond: integrating molecular'omics' for microbial community profiling. Nature Reviews Microbiology 136, 360-372.
- Fütterer, Angelov, Liesegang, Gottschalk, et al. 2004. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proceedings of the National Academy of Sciences of the United States of America 10124, 9091-9096.
- Handelsman 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiology and molecular biology reviews 684, 669-685.
- Kawashima, Amano, Koike, Makino, et al. 2000. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proceedings of the National Academy of Sciences 9726, 14257-14262.
- Kazanov, Vitreschak and Gelfand 2007. Abundance and functional diversity of riboswitches in microbial communities. BMC genomics 81, 1.
- Mandal, Boese, Barrick, Winkler et al. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 1135, 577-586.
- Mandal and Breaker 2004. Gene regulation by riboswitches. Nature Reviews Molecular Cell Biology 56, 451-463.
- Marsh 1998. Coenzyme B12 (cobalamin)-dependent enzymes. Essays in biochemistry 34139-154.
- Mukherjee and Sengupta 2015. Riboswitch Scanner: an efficient pHMM-based web-server to detect riboswitches in genomic sequences. Bioinformaticsbtv640.
- Nicol and Schleper 2006. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends in microbiology 145, 207-212.
- Preston, Wu, Molinski and DeLong 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proceedings of the National Academy of Sciences 9313, 6241-6246.
- Rodionov, Vitreschak, Mironov and Gelfand 2002. Comparative genomics of thiamin biosynthesis in procaryotes New genes and regulatory mechanisms. Journal of Biological chemistry 27750, 48949-48959.
- Rother, Rother, Puton and Bujnicki 2011. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic acids research 3910, 4007-4022.
- Ruepp, Graml, Santos-Martinez, Koretke, et al. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 4076803, 508-513.
- Serganov, Polonskaia, Phan, Breaker et al. 2006. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 4417097, 1167-1171.
- Strobel and Cochrane 2007. RNA catalysis: ribozymes, ribosomes, and riboswitches. Current opinion in chemical biology 116, 636-643.
- Sudarsan, Barrick and BREAKER 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. Rna 96, 644-647.
- Thomas, Gilbert and Meyer 2012. Metagenomics-a guide from sampling to data analysis. Microbial informatics and experimentation 21, 1.
- Vitreschak, Rodionov, Mironov and Gelfand 2004. Riboswitches: the oldest mechanism for the regulation of gene expression? TRENDS in Genetics 201, 44-50.
- Winkler and Breaker 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59487-517.
- Yadav, Swati and Chandrasekharan 2015. Thiamine Pyrophosphate Riboswitch in Some Representative Plant Species: A Bioinformatics Study. Journal of Computational Biology 22, 1-9.