Research Article - Biomedical Research (2018) Volume 29, Issue 9
Exosomal natural antisense transcripts in culture supernatants released from colorectal cancer SW480 cells
Mitsuru Chiba*
Department of Bioscience and Laboratory Medicine, Graduate School of Health Sciences, Hirosaki University, 66-1 Hon-Cho, Hirosaki, Aomori, Japan
- *Corresponding Author:
- Mitsuru Chiba
Department of Bioscience and Laboratory Medicine
Graduate School of Health Sciences
Hirosaki University, 66-1 Hon-Cho
Hirosaki, Aomori, Japan, Chiba, 1919
Accepted on March 09, 2018
DOI: 10.4066/biomedicalresearch.29-18-272
Visit for more related articles at Biomedical ResearchAbstract
Introduction: Exosomes are extracellular vesicles, which are released from most cells, involved in cell-tocell communication. Exosomes contain membrane and cytosolic components such as proteins, lipids, messenger Ribonucleic Acids (mRNAs), and microRNAs. However, the existence of other types of RNAs in these exosomes is unknown.
Methods: In the present study, the presence of Natural Antisense Transcripts (NATs) transcribed from reverse strand deoxynucleic acids of genes in exosomes released from colorectal cancer SW480 cells were investigated using a sense/antisense custom microarray.
Results and Discussion: When signal intensities with values of more than 100 were selected, 6,218 NATs were detected in exosomes released from SW480 cells. Among them, NATs transcribed from transcription factor 7-like 1 genes, which had the lowest signal intensities, were detected using strandspecific reverse transcription-polymerase chain reaction. Thus, a large number of NATs as well as mRNAs and microRNAs are present in exosomes and are likely to be delivered to other cells.
Keywords
Exosomes, Extracellular vesicle, Natural antisense transcript, Microarray, SW480.
Introduction
Natural Antisense Transcripts (NATs) are Ribonucleic Acids (RNAs) that are transcribed from the opposite deoxynucleic acids (DNAs) strand of a gene and overlap in part with sense transcripts. NATs have been discovered using the analyses of human and mouse genome sequence project and full-length complementary DNAs (cDNAs). Kiyosawa et al. classified sense/antisense transcript pairs according to overlap patterns and protein or non-protein-coding patterns [1]. The functional annotation of the mammalian genome transcriptome sequencing consortium identified NATs for more than 70% of transcription units, most of which represent non-protein-coding RNAs [2]. A number of mechanisms have been proposed for NATs-mediated regulation of sense transcripts, including messenger RNAs (mRNAs). NATs localized in the nucleus are involved in transcriptional interference regulation, chromatin induction and DNA epigenetic changes, genomic imprinting, X-chromosome inactivation and alternative splicing [3-7]. NATs in the cytoplasm are involved in mRNA stability regulation, translational inhibition and endogenous small interfering RNA formation [8-10].
Recently, some researchers have investigated the role of NATs in several diseases. Differential NAT expressions were observed between cancerous tissues and the corresponding non-cancerous tissues obtained from colorectal and breast cancers [11,12]. In addition, Matsui et al. discovered that NATs of inducible nitric oxide synthase (iNOS), an important gene in inflammatory diseases, increased the stability of iNOS mRNA [8]. Furthermore, Faghihi et al. reported that NATs of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) increased the stability of BACE1 mRNAs by forming RNA duplex [13]. Thus, it is suggested that NATs are associated with the development and progression of several diseases.
Exosomes are extracellular vesicles (approximately 40-200 nm in diameter) of endocytic origin released from cells and contain proteins, lipids, and nucleic acids (DNAs and RNAs) derived from the cell [14]. Valadi et al. discovered mRNAs and microRNAs inside exosomes released from mast cells, showing for the first time that mRNAs and microRNAs could be delivered from one cell to other cells [15]. ExoCarta (http://www.exocarta.org/) is a database of exosomal components identified by both published and unpublished studies [16]. At this time, proteins (4,563 entities), mRNAs (1,639 entities), and microRNAs (764 entities), and lipids (194 entities) have been registered in the database (release date: May 29, 2012). It was first identified NATs transcribed from leucine-rich repeatcontaining 24 (LRRC24), MDM2 proto-oncogene E3 ubiquitin protein ligase (MDM2), and cyclin-dependent kinase inhibitor 1A (CDKN1A) genes in exosomes released from three human colorectal cancer cells [17]. However, the types of exosomal NATs are unknown till date.
In the present study, the types of exosomal NATs were investigated using human colorectal cancer SW480 cells and a custom microarray containing human sense/antisense probes for approximately 21,000 genes.
Materials and Methods
Cell cultures
The human colorectal cancer cell line SW480 (CCL-228) was purchased from the American Type Culture Collection (ATCC, Manassas, VA). These cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Wako, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS; Life Technologies, Carlsbad, CA), 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were cultured at 37°C in a humidified atmosphere of 95% air/5% CO2.
Collection of exosomes from culture supernatants
SW480 cells were plated on collagen-coated 10-cm dishes at 1 × 106 cells per dish with the culture media described above. After 72 h, the culture medium was discarded, and the cells were washed three times in a serum-free culture medium. Then, 10 ml serum-free culture medium was added to each dish, and the cells were cultured for 48 h. Cell cultures from each dish were collected and subjected to a sequence of centrifugation steps. Cell cultures were first centrifuged at 300 × g at 4°C for 3 min to remove floating cells. These supernatants were then centrifuged at 2,000 × g at 4°C for 15 min and collected in new tubes. In addition, culture supernatants were centrifuged at 12,000 × g at 4°C for 35 min to remove cellular debris, and these supernatants were collected in new tubes. Supernatants were then filtered through a 0.22 μm filter. These filtrates were ultra-centrifuged at 120,000 × g at 4°C for 70 min to collect exosomes using the Optima TLX Ultracentrifuge (Beckman Coulter, Brea, CA). Exosome pellets were washed in Dulbecco’s phosphate buffered saline (D-PBS) and further ultra-centrifuged at 120,000 × g at 4°C for 70 min. Finally, exosome pellets were re-suspended in D-PBS.
RNA extraction
Isogen II (NipponGene, Tokyo, Japan) was used to isolate exosomal RNAs from exosome pellets. Exosomal RNAs were stored at -80°C until further use. The concentrations of RNAs extracted were examined using the Quant-iT RiboGreen RNA Reagent and Kit (ThermoFisher Scientific, DE, USA), according to the manufacturer’s instructions. In addition, the size of the exosomal RNA was confirmed using the Agilent 2100 Bioanalyzer and Agilent RNA 6000 Pico Kit (Agilent Technologies, Foster City, CA), according to the manufacturer’s instructions.
Microarray analysis
Exosomal RNAs (1 ng) derived from SW480 cells were amplified using Ovation Pico WTA System V2 (NuGEN Technologies, San Carlos, CA). Amplified exosomal RNAs were purified using the QIAquick Nucleotide Removal Kit (Qiagen, Valencia, CA). Cyanine 3 (Cy3)-labelled cDNAs were synthesized using the Agilent Genomic DNA Enzymatic Labeling Kit (Agilent Technologies), according to the manufacturer’s instructions. Cy3-labeled cDNAs were purified using the QIAquick Nucleotide Removal Kit (Qiagen). The concentration and quality of Cy3-labeled cDNAs were measured using the Agilent 2100 Bioanalyzer. Agilent 44 K × 4 human custom microarray slides (Agilent eArray Design ID=19052, produced by Tsukuba GeneTech Laboratory, Ibaraki, Japan) were hybridized with Cy3-labeled cDNA (5 μg) in hybridization solution and blocking agent (Agilent Technologies) at 65°C for 24 h using a hybridization oven. Microarray slides were washed in wash Buffer 1 and 2 (Agilent Technologies) including 0.005% Triton X-102. Washed microarray slides were dried using nitrogen gas. Cy3 fluorescence images of array slides were obtained using a DNA microarray scanner (Agilent Technologies) and were processed using Feature Extraction version 8.1 software based on instructions from Agilent Technologies.
Strand-specific reverse transcription polymerase chain reaction (RT-PCR)
First-strand cDNAs from NATs were synthesized using exosomal RNAs (1 ng), RT primer, and AMV Reverse Transcriptase (Promega, Madison, WI) according to the procedure recommended by Promega. The mixtures were then incubated at 50°C for 60 min. The resulting cDNAs were incubated at 99°C for 5 min and at 37°C for 60 min with ribonuclease A to digest RNAs. The first-strand cDNAs were subjected to ordinary PCR using forward and reverse primers.
The detection of NATs by PCR was performed using firststrand cDNAs, FastStart Universal SYBR Green Master (Roche Diagnostics, Basel, Switzerland), and primer pairs. PCR analyses were performed using the StepOne Plus Real- Time PCR system (RT-PCR; Life Technologies, Carlsbad, CA, USA) for 1 min at 95°C followed by 40 cycles each of 95°C for 15 s and 60°C for 60 s, following the manufacturer’s instructions. The PCR products obtained above were electrophoresed using 4% agarose gels. The detection of amplified fragments was achieved by ethidium bromide staining using the ChemiDoc XRS (Bio-Rad) and the Quantity One software (Bio-Rad).
Results and Discussion
Detection of exosomal RNAs released from SW480 cells
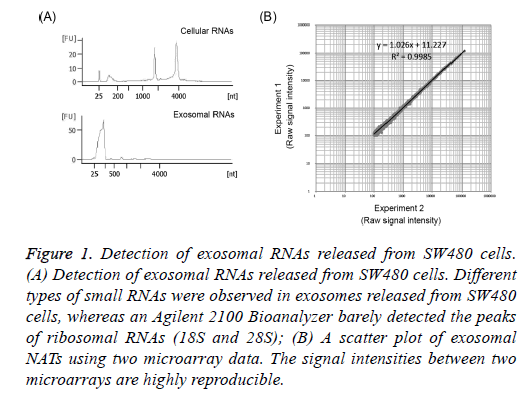
To confirm that exosomes released from SW480 cells had been collected, exosomal RNAs were extracted from the precipitation of cells or exosomes obtained from supernatants by ultracentrifugation using Isogen II reagent. Exosomal and/or cellular RNAs obtained were electrophoresed using the Agilent 2100 Bioanalyzer. The 18S and 28S ribosomal RNAs were detected from the RNAs of SW480 cells (Figure 1A). On the other hand, their ribosomal RNAs were barely detected in exosomal RNAs and small RNAs were detected in the size range of 25-200 nucleotides (nt) in these exosomal RNAs (Figure 1A). It was reported that several small RNAs were observed in exosomes [17]. This indicates that exosomes released from the supernatants of SW480 cells were collected in the present study.
Figure 1: Detection of exosomal RNAs released from SW480 cells. (A) Detection of exosomal RNAs released from SW480 cells. Different types of small RNAs were observed in exosomes released from SW480 cells, whereas an Agilent 2100 Bioanalyzer barely detected the peaks of ribosomal RNAs (18S and 28S); (B) A scatter plot of exosomal NATs using two microarray data. The signal intensities between two microarrays are highly reproducible.
Expression profiles of exosomal NATs
Many researchers have shown that exosomes released from cells include microRNAs that are delivered to other cells [15,17-20]. Exosomal RNAs other than microRNAs have also been reported. Valadi et al. reported that exosomes released from mast cells included mRNA [15]. In addition, Lambertz et al. demonstrated that transfer RNAs (tRNAs) were detected in exosomes released from Leishmania [21]. In an earlier study, it was found that NATs of MDM2 and CDKN1A genes in exosomes released from human colorectal cancer cells [17]. However, the types of NATs in exosomes are unknown till date. In the present study, the types of exosomal NATs were investigated using human colorectal cancer SW480 cells and a custom microarray containing human sense/antisense probes.
A high peak of small RNAs was detected in exosomal RNAs from SW480 cells (Figure 1A). Because NATs include RNAs of longer lengths, it is necessary to amplify exosomal RNAs for the microarray analysis of NATs. Dafforn et al. reported small amounts of RNA amplification method, Ribo-SPIA, which generates micrograms of labelled cDNAs from 5 ng total RNAs in 1 day for microarray analysis [22]. Here I identified NATs using amplified exosomal RNAs by Ribo- SPIA method and a sense/antisense custom microarray. In addition, the reproducibility of this experiment was investigated. When signal intensities with a value of more than 100 were selected, 6,218 NATs were detected in exosomes released from SW480 cells (Tables 1 and 2). The amplified exosomal RNAs prepared using the Ribo-SPIA procedure indicate an accurate amplification of gene expressions (R2=0.9985) (Figure 1B). This result indicates that a large number of NATs were observed in exosomes released from SW480 cells. This data (GEO accession: GSE68979) is registered in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/).
| Accession number | Gene symbol | Description | Signal value (g processed signal) |
|---|---|---|---|
| XM_001127227.1 | LOC728331 | PREDICTED: Homo sapiens hypothetical protein LOC728331 (LOC728331), mRNA | 119623.8 |
| NM_033301.1 | RPL8 | Homo sapiens ribosomal protein L8 (RPL8), transcript variant 2, mRNA | 86993.31 |
| NM_001015.3 | RPS11 | Homo sapiens ribosomal protein S11 (RPS11), mRNA | 78607.87 |
| NM_004559.2 | YBX1 | Homo sapiens Y box binding protein 1 (YBX1), mRNA | 75369.76 |
| XM_001126335.1 | LOC727826 | PREDICTED: Homo sapiens similar to ribosomal protein S11 (LOC727826), mRNA | 73983.69 |
| NR_002202.1 | FTHL7 | Homo sapiens ferritin, heavy polypeptide-like 7 (FTHL7) on chromosome 13 | 73893.05 |
| XR_016327.1 | LOC338870 | PREDICTED: Homo sapiens similar to ribosomal protein S12 (LOC338870), mRNA | 69479.47 |
| XR_015536.1 | LOC731599 | PREDICTED: Homo sapiens hypothetical protein LOC731599 (LOC731599), mRNA | 68422.79 |
| NM_000973.3 | RPL8 | Homo sapiens ribosomal protein L8 (RPL8), transcript variant 1, mRNA | 68120.92 |
| NM_006401.1 | ANP32B | Homo sapiens acidic (leucine-rich) nuclear phosphoprotein 32 family, member B (ANP32B), mRNA | 67702.16 |
Table 1. Top 10 (of the 6,218 indicated) natural antisense transcripts in exosomes released from SW480 cells.
| Accession number | Gene symbol | Description | Signal value (g processed signal) |
|---|---|---|---|
| NM_014028.3 | OSTM1 | Homo sapiens osteopetrosis associated transmembrane protein 1 (OSTM1), mRNA | 101.94 |
| NM_006184.3 | NUCB1 | Homo sapiens nucleobindin 1 (NUCB1), mRNA | 101.8 |
| XM_001127824.1 | LOC728574 | PREDICTED: Homo sapiens similar to retinitis pigmentosa GTPase regulator isoform C (LOC728574), mRNA | 101.71 |
| NM_022727.4 | HTF9C | Homo sapiens HpaII tiny fragments locus 9C (HTF9C), transcript variant 1, mRNA | 101.67 |
| NM_001040445.1 | ASB1 | Homo sapiens ankyrin repeat and SOCS box-containing 1 (ASB1), mRNA | 101.56 |
| NM_147152.1 | ITSN2 | Homo sapiens intersectin 2 (ITSN2), transcript variant 2, mRNA | 101.43 |
| NM_152682.1 | RWDD4A | Homo sapiens RWD domain containing 4A (RWDD4A), mRNA | 101.38 |
| NM_000292.1 | PHKA2 | Homo sapiens phosphorylase kinase, alpha 2 (liver) (PHKA2), mRNA | 101.19 |
| NM_031283.1 | TCF7L1 | Homo sapiens transcription factor 7-like 1 (T-cell specific, HMG-box) (TCF7L1), mRNA | 101.19 |
| NM_002269.2 | KPNA5 | Homo sapiens karyopherin alpha 5 (importin alpha 6) (KPNA5), mRNA | 100.5 |
Table 2. Bottom 10 (of the 6,218 indicated) natural antisense transcripts in exosomes released from SW480 cells.
Detection of exosomal NATs of TCF7L1 gene
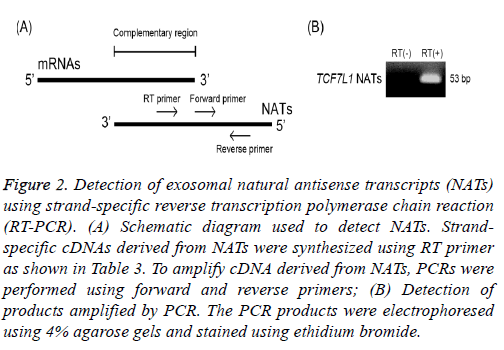
To validate these exosomal NATs as detected above, NATs transcribed from transcription factor 7-like 1 (TCF7L1) gene, which were found to have the lowest signal intensities, were investigated using strand-specific RT-PCR. It is difficult to specifically detect NATs by the reaction of reverse transcription using oligo dT primer or a random primer because a part of NATs has the complementary sequence within a region of mRNAs (Figure 2A). Therefore, I synthesized strand-specific cDNA derived from NATs of TCF7L1 using RT primer (Table 3). The obtained cDNAs were subjected to RT-PCR using a SYBR green reagent and amplified. These amplicons were electrophoresed using 4% agarose gel and stained with ethidium bromide. Band of TCF7L1 NATs detected using ultraviolet irradiation were 53 base pair (bp), respectively (Figure 2B). This indicates that NATs of TCF7L1 are present in exosomes released from SW480 cells, suggesting that other higher-intensity NATs detected using microarray analysis are also present in these exosomes.
| Primer name | Accession no. | Sequence (5′-3′) | Size (nt) | Amplicon size (bp) |
|---|---|---|---|---|
| TCF7L1 RT primer | NM_031283.1 | AGCTTTCAGTGGAGAAAG | 18 | - |
| TCF7L1 forward primer | GGTCCTCTGAAATGGTTTC | 19 | 53 | |
| TCF7L1 reverse primer | CACCTTGAGTCCCTTTAAATG | 21 |
Table 3. Primers for strand-specific reverse transcription polymerase chain reaction.
Figure 2: Detection of exosomal natural antisense transcripts (NATs) using strand-specific reverse transcription polymerase chain reaction (RT-PCR). (A) Schematic diagram used to detect NATs. Strandspecific cDNAs derived from NATs were synthesized using RT primer as shown in Table 3. To amplify cDNA derived from NATs, PCRs were performed using forward and reverse primers; (B) Detection of products amplified by PCR. The PCR products were electrophoresed using 4% agarose gels and stained using ethidium bromide.
Recently, the analyses of exosomal components were performed using next-generation sequencers. Vojtech et al. identified exosomal piwi-RNAs (small RNAs (26-31 nt) expressed primarily in germline cells) and Y RNAs (small RNAs (83-112 nt) involved in alternative splicing and regulating translation) from human semen exosomes using the HiSeq 2000 sequencing system by Illumina [23]. Their exosomal RNAs may be also delivered to other cells and functions in target cells. It is necessary to elucidate whether a number of NATs identified in the present study are delivered to other cells and their functions in target cells. In conclusion, a large number of NATs are present in exosomes released from colorectal cancer cells, suggesting that these NATs are also delivered to other cells.
Conclusion
A large number of NATs are present in exosomes released from SW480 cells.
Competing Interests
No competing interests exist.
Acknowledgement
This work was supported in part by a Hirosaki University Institutional Research Grant for Young Scientists, JSPS KAKENHI (no. 23790613, 25670264, and 17K19779), a grant from the Suzuken Memorial Foundation (no. 11-076), a grant from the Takeda Science Foundation, and a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT).
References
- Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res 2003; 13: 1324-1334.
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science 2005; 309: 1564-1566.
- Shearwin KE, Callen BP, Egan JB: Transcriptional interference-a crash course. Trends Genet 2005; 21: 339-345.
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008; 451: 202-206.
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002; 415: 810-813.
- Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 2008; 135: 227-235.
- Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res 1997; 25: 4296-4300.
- Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology 2008; 47: 686-697.
- Hatzoglou A, Deshayes F, Madry C, Lapree G, Castanas E, Tsapis A. Natural antisense RNA inhibits the expression of BCMA, a tumour necrosis factor receptor homologue. BMC Mol Biol 2002; 3: 4.
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005; 123: 1279-1291.
- Kohno K, Chiba M, Murata S, Pak S, Nagai K, Yamamoto M, Yanagisawa K, Kobayashi A, Yasue H, Ohkohchi N. Identification of natural antisense transcripts involved in human colorectal cancer development. Int J Oncol 2010; 37: 1425-1432.
- Grigoriadis A, Oliver GR, Tanney A, Kendrick H, Smalley MJ, Jat P, Neville AM. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genomics 2009; 10: 324.
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 2008; 14: 723-730.
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010; 73: 1907-1920.
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654-659.
- Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles 2012; 1.
- Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep 2012; 28: 1551-1558.
- Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem 2014; 289: 22258-22267.
- Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer 2011; 10: 117.
- Cho JA, Yeo DJ, Son HY, Kim HW, Jung DS, Ko JK, Koh JS, Kim YN, Kim CW. Exosomes: a new delivery system for tumor antigens in cancer immunotherapy. Int J Cancer 2005; 114: 613-622.
- Lambertz U, Oviedo Ovando ME, Vasconcelos E, Unrau PJ, Myler PJ, Reiner NE. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genomics 2015; 16: 151.
- Dafforn A, Chen P, Deng G. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques 2004; 37: 854-857.
- Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 2014; 42: 7290-7304.