Research Article - Biomedical Research (2018) Volume 29, Issue 21
Elevated SAP/SLAM expression in hepatitis B vaccine responders, but not in non-responders among chronic hepatitis C patients.
Jian Fu1, Xuejiao Pan1, Jun Yao1, Shuyun Xie1, Huakun Lv1, Zhifang Wang2,3* and Chonggao Hu3*
1Department of Immunization Programme, Zhejiang Provincial Center for Disease Control and Prevention, 3399 Binsheng Road, Hangzhou, China
2Department of Environmental and Occupational Health, Zhejiang Provincial Center for Disease Control and Prevention, 3399 Binsheng Road, Hangzhou, China
3Key Medical Research Center, Zhejiang Provincial Center for Disease Control and Prevention, 3399 Binsheng Road, Hangzhou, China
- *Corresponding Author:
- Zhifang Wang
Department of Environmental and Occupational Health
Zhejiang Provincial Center for Disease Control and Prevention
3399 Binsheng Road, Hangzhou, China
Chonggao Hu
Key Medical Research Center
Zhejiang Provincial Center for Disease Control and Prevention
3399 Binsheng Road, Hangzhou, China
Accepted on July 13, 2018
DOI: 10.4066/biomedicalresearch.29-18-819
Visit for more related articles at Biomedical ResearchAbstract
About 5%-10% of vaccinated individuals fail to produce a protective level of anti-hepatitis B surface antibody (anti-HBs) though hepatitis B vaccines show excellent immunogenicity in the general population. The percentage of humoral immunity failure to hepatitis B vaccination is about 30% in individuals infected with hepatitis C virus (HCV). However, mechanisms on the higher non-responsive rate in HCV patients are still unknown. Our study was conducted to describe the potential mechanisms of the Singling Lymphocytic Activation Molecule (SLAM) and SLAM-Associated Protein (SAP) between T cells and B cells borders during humoral immunity response. Based on anti-HBs Ab titers post immunization, we designated three groups: healthy controls (HC), vaccine responders in HCV-infected patients (VR), and vaccine non-responders in HCV-infected patients (VNR), respectively. Protein and mRNA expression levels of SAP and SLAM were determined among three groups at two time points, pre-vaccination and post-vaccination, respectively. For VR and HC, mRNA and protein expression levels of SAP and SLAM were elevated post-vaccination. However, these changes were not seen in VNR. Increased expression levels of SAP and SLAM may be associated with the development of a protective anti-HBs Ab response to hepatitis B vaccination. Therefore, modulation of SAP and SLAM may be considered as a strategy to improve vaccination for HCV-infected patients.
Keywords
Humoral immunity, Hepatitis B vaccine, Hepatitis C virus, SAP, SLAM.
Introduction
Globally, Hepatitis B Virus (HBV) infection affects more than one third of the population, with an incidence rate of 1.26 million deaths per year [1]. Immunization with hepatitis B vaccine containing the major surface antigen of HBV (HBs Ag) is considered as the economic, efficient and safe strategy to prevent and control this disease. Patients infected with hepatitis C virus (HCV) are suggested to be immunized with hepatitis B vaccine due to co-infections with HBV and HCV can increase severity of hepatitis and serious complications. Though hepatitis B vaccine show excellent immunogenicity in the general population with normal immune status, approximately 30% of chronic hepatitis C patients fail to develop protective hepatitis B surface antibody (defined as anti-HBs titers are less than 10 mIU/ml) after a series of standard vaccination schedule (involving 3 doses of 20 μg HBs Ag vaccine at 0, 1, 6 months) with hepatitis B vaccine [2-4].
Many factors have been recognized as the potential causes for failure to induce a protective anti-HBs response to hepatitis B vaccination, such as host-related factors (increasing age, obesity, smoking cigarettes, as well as male gender) and genetic factors (specific human leukocyte antigen genes) [5,6]. Recent studies show that the importance of signaling lymphocytic activation molecule (SLAM)-associate protein (SAP) as an intracellular signal molecular in humoral immunity [7,8]. SAP-deficient mice lack long-term humoral immunity after immunization by their paucity of antigen (Ag)- specific memory B cells and long-lived Ab-secreting plasma cells [9-12]. SAP-mutated patients in X-linked lymphoproliferative disease character a dysregulated immune response [13]. Although defects in several immune cell types associated with SAP were found to relate with the humoral immunity response, these studies were usually implemented on the patients and the mice models. The patients with immunodeficiency disorders may complicate the interpretation of the data; the immune system in mice distinguishes from that in human beings due to different species. In addition, analysis on the association SAP/SLAM family adhesions and immune response to hepatitis B vaccine is crucial to shed light on the underlying mechanisms on vaccination failure. However, to date, the underlying mechanisms on how SAP regulates lymphocyte functions are not well understood in humans with normal immune status. In this study, we undertook to address this issue between hepatitis B vaccine responders and nonresponders in healthy individuals and chronic hepatitis C patients.
Materials and Methods
Vaccine schedule and study subjects
Between February 2013 and June 2016, vaccines were voluntarily immunized with 3-dose 20 μg recombinant Chinese hamster ovary cell HB vaccine (200802A21, North China Pharmaceutical Group Corp GeneTech Biotechnology Development Co., Ltd.) at 0, 1 and 6 months. Peripheral venous blood (10 ml) was collected in plastic tubes (BD Biosciences) under sterile condition at two time points: before the first dose vaccine (T0) and 4 weeks after 3 doses vaccine (T1). Anti-HBs titers were measured by enzyme-linked immunosorbent assay (ELISA) using a commercial kit (ABIN509703, antibodies-online Inc.).
Vaccine non-responders were defined as participants whose anti-HBs titers were lower than 10 mIU/ml. Vaccine responders were designated as subjects whose anti-HBs titers were greater than 10 mIU/ml; In this study, total of 8 vaccine responders without hepatitis C virus infection were selected as healthy controls (HC); 17 participants infected with hepatitis C virus were enrolled, including 8 vaccine responders (VR) and 9 vaccine non-responders (VNR). Characteristics of study participants were shown in Table 1.
| HCV RNA | Anti-HBs titers (mIU/ml) | ||||
|---|---|---|---|---|---|
| Groups | Age (Mean ± SD) | Gender | (log10 copies/ml) | T0 | T1 |
| HC | 30.9 ± 15.6 y | M=11; F=9 | NA | 1.5 ± 0.4 | 205.3 ± 57.0 |
| VR | 48.1 ± 18.4 y | M=6; F=10 | 6.1 ± 3.2 | 0.7 ± 0.2 | 197.2 ± 34.3 |
| VNR | 60.5 ± 20.4 y | M=11; F=7 | 5.2 ± 2.1 | 1.2 ± 0.7 | 8.5 ± 0.4 |
SD: Standard Deviation; NA: Not Applicable; HC: Healthy Controls; VR: Vaccine Responders among chronic hepatitis C patients; VNR: Vaccine Non-Responders among chronic hepatitis C patients.
Table 1. Characteristics of study participants and their anti-HBs responses.
RNA preparation, reverse transcription and qRTPCR
Total RNA from 106 PBMC was extracted using TRI Reagent RNA Isolation Kit (T9424, Sigma Molecular Research Center, Inc.) following the manufacturer’s instructions. The concentration and purity of the RNA was quantified by a spectrophotometer (NanoDrop 8000; Thermo Fisher Scientific) using A260/A280 ratio and A230/A260 ratio. Samples with RNA concentration (A260/A280 ≥ 1.7 ng/μl) and purity (1.5 ng/μl ≤ A230/A260 ≤ 2.4 ng/μl) were chosen for further analysis.
Extracted RNA (3.0 μg) from each sample was used as template for cDNA generation with 3.6 μM oligo-dT primers in the synthesis solution (20 μL), using Thermo Scientific Verso cDNA Synthesis kit (AB-1453/A). Reactions were incubated at 70°C for 5 min and then place immediately on ice. In addition, a total of 40 units of RNaseOUT Recombinant RNase inhibitor (Verso cDNA Kit, AB1453A, Thermo Fisher Scientific) and 14 mM dithiothreitol were added to the mixed reaction. Next, reverse transcriptase (1 μL) was added and the reactions started at 42°C for 30 min and 95°C for 2 min to inactivate reverse transcriptase. cDNA samples were diluted to a RNA equivalent of 1 ng/μL with DEPC-treated water for downstream processing. Each sample in the test was assayed in triplicate.
We performed quantitative real-time quantitative PCR assays (qRT-PCR) in Bio-Rad DNA Engine Opticon 2 System instrument (Bio-Rad Laboratories, Hercules, CA). Reactions (20 μL/well) were performed in skirted PCR plates consisting of 3 μL cDNA sample, 10 μL iQ SYBR green super mix (170-8882; Bio-Rad Laboratories) and 300 nM the following primers: SLAM-forward: 5’-AGC AGG TCT CCA CTC CAGAA-3’; SLAM-reverse: 5’-GCT CAC GGT GCA GAT GTAGA-3’; SAP-forward:5’-AAC TGG GAG TCA GGT GGTTG-3’; SAP-reverse: 5’-GCT GTC CCT CAG CAA ATAGC-3’). Cycling conditions for each plate included a 95°C for incubation for 2 min followed by 40 cycles of 95°C for 10 s and 55°C for 30 s. Melting curve analysis was performed between 55°C and 95°C. We used β-actin housekeeping gene as the internal control under the same PCR conditions described for SLAM and SAP.
Protein extraction and Western blotting analysis
Peripheral Blood Mononuclear Cells (PBMC) was isolated by density gradient centrifugation on Ficoll-Paque (Sigma, USA). We extracted total cell protein from each sample by lysing cell in the cold radioimmuno-precipitation assay buffer in the presence of protease inhibitors. The amount of protein was determined by Coomassie Protein Assay Reagent Kit (23200, Thermo Fisher Scientific) according to the manufacturer’s instructions. Protein was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis in 10% polyacrylamide gels and blotted onto PVDF membrane. Blots were blocked in PBS with 0.5% skim milk for 2 h at room temperature and incubated with primary and secondary antibodies. Monoclonal antibodies against CD150 (sc-166939, Santa Cruz Biotechnology) and SH2D1A (sc-398118, Santa Cruz Biotechnology) were used at a dilution of 1:500 and 1:200, respectively. Secondary antibody (sc-2005, Santa Cruz Biotechnology) was used at dilution of 1:1000. The super signal chemiluminescent substrate detection kit (34094, Thermo Fisher Scientific) and ECL film (Kodak, China) were used to visualize the presence of specific protein in the PVDF blots. Immunoreactive bands were quantified by volume densitometry using Image J Basics Version 1.49 (http:// rsb.info.nih.gov/ij/download.html) and normalized to β-actin (sc-47778, Santa Cruz Biotechnology). All data represent the results of at least three independent assays.
Statistical analysis
SAP/SLAM mRNA and protein levels were quantified in each study participant at two time points: T0 and T1. Results were expressed as ratios and normalized by the internal control (β- actin). Statistical analysis was performed using SPSS version 23.0 (SPSS Inc. Chicago, IL, USA). Statistically significant differences were determined using paired t-tests. A P value less than 0.05 were considered as a statistical significance.
Ethical statement
Written informed consent was obtained from each participant. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by Zhejiang Provincial Center for Disease Control and Prevention Ethics Committee.
Results
Increased SAP/SLAM mRNA levels in producing protective anti-HBs response among HC and VR, but not among VNR
SAP/SLAM mRNA levels were determined by qRT-PCR before and after vaccination with recombinant hepatitis B vaccine (Table 2). Compared with the SAP mRNA levels at T0, they were significantly elevated for persons developing a protective antibody response among HC and VR at T1. However, the similarly increased SAP mRNA levels were not observed for VNR. Post immunization, the relative SLAM mRNA levels were significantly increased for HC and VR, but not for VNR.
| mRNA | Groups | T0 | T1 | P value |
|---|---|---|---|---|
| SAP | HC | 10.7 ± 4.19 | 14.48 ± 6.35 | 0.001* |
| VR | 7.98 ± 5.24 | 10.08 ± 4.28 | 0.005* | |
| VNR | 10.20 ± 3.44 | 11.15 ± 5.4 | 0.186 | |
| SLAM | HC | 7.86 ± 5.02 | 10.53 ± 6.32 | 0.034* |
| VR | 6.69 ± 5.32 | 12.24 ± 6.78 | 0.039* | |
| VNR | 7.89 ± 4.81 | 6.92 ± 5.89 | 0.981 | |
| mRNA: message RNA; HC: Healthy Controls; VR: Vaccine Responders; VNR: Vaccine Non-Responders; *P<0.05. | ||||
Table 2. mRNA levels in HC, VR and VNR to hepatitis B vaccine.
SAP mRNA levels are positively correlated with SLAM in three groups
SAP is a SRC homology 2 (SH2) domain-containing molecular. In vivo, expression of SAP is regulated following immune activation [14]. After vaccination, SAP functions as a transduced signal between T cells and B cells by binding of the cytoplasmic domain of SLAM. To establish the relationship between SAP and SLAM mRNA levels in three different groups before and after vaccination with recombined hepatitis B vaccine, we calculated the correlation coefficients by correlation analysis (Table 3). It was shown that SAP mRNA levels in peripheral blood among HC, VR, and VNR were significantly positively related with SLAM mRNA levels.
| Groups | Time points | Correlation coefficient | P value |
|---|---|---|---|
| HC | T0 | 0.81 | 0.0006** |
| T1 | 0.83 | 0.0086* | |
| VR | T0 | 0.76 | 0.0001** |
| T1 | 0.91 | 0.03* | |
| VNR | T0 | 0.79 | 0.01* |
| T1 | 0.9 | 0.0001** | |
| HC: Healthy Controls; VR: Vaccine Responders; VNR: Vaccine Non-Responders; *P<0.05; **P<0.001. | |||
Table 3. Correlation analysis between SAP and SLAM based on mRNA levels.
Increased SAP/SLAM protein levels post vaccination for HC and VR but not for VNR
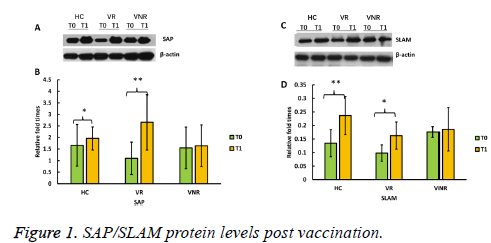
To show whether SAP/SLAM protein levels increased after vaccination with recombined hepatitis B vaccine, we further determined each protein expression level by Western blot (Figure 1).
The results showed that SAP protein levels were significantly higher post vaccination (T1) than pre-vaccination (T0) among HC and VR, however, SAP protein level was not increased among VNR post immunization (T1) (Figures 1A and Figure 1B).
Similarly, SLAM proteins levels were significantly enhanced post vaccination (T1) among HC and VR not among VNR, compared with the level before the day of vaccination (T0) (Figures 1C and Figure 1D).
Discussion
To our knowledge, this is the first time to determine, postimmunization with recombined hepatitis B vaccine, the expression levels of both SAP and SLAM were increased among HC and VR who had a protective anti-HBs response but not among VNR who failed to produce a protective humoral immunity response.
Post immunization with recombined hepatitis B vaccine, CD4 T cells interact with antigen-presenting cells, proliferate, activate, and migrate toward the T-B cell border in germinal centers (GCs). This cooperation between T cells and B cells require direct physical interactions, which induce maturation of HBs antigen-specific B cells and production of long-lived of plasma cell and generate long-term anti-HBs.
For humoral immunity response, SAP family adaptors have been found to be expressed in B cells and T cells [12,15]. Previous studies in SAP-deficient mice show that, SAP play important roles for T cell functions and B cells differentiation, maturation and antibody production [16,17]. Therefore, SAPdeficient mice have flaws in humoral immunity to reproduce the antibody by severely decreasing to form GCs, losing of long-term antibody-secreting cells and reducing serum IgG. In this study, we show in the unique design of recombined hepatitis B vaccination-induced antibody responses in chronic hepatitis C virus-infected patients and healthy controls, SAP expression levels of vaccine responders were quantitatively superior to those of vaccine non-responders despite being similar levels before vaccination in different research groups. This result is in agreement with existing evidence that SAP is needed to mediate signal transduction as cell adhesions to produce humoral immune response [18,19]. This result is also agreement with the studies in SAP-deficient CD4 T cells in vitro and in SAP-deficient mice in vivo [20,21]. These phenomena imply that SAP up-regulation is necessary for T-B cell adhesions to produce efficient antigen-specific antibody post immunization.
SLAM family of receptors express on the surface of a wide range of immune cells, including CD4 T cells, CD8 T cells, macrophages, dendritic cells, natural killer cells and B cells.
There is accumulating evidence that SLAM family receptors are necessary to form stable conjugates between T cells and antigen-specific B cells [18,21]. In the present study, we also observed that mRNA expression levels of SLAM were up regulated post-immunization among HC and VR but not among VNR. This phenomenon implies that SLAM is also crucial for a protective humoral immunity to have efficient anti-HBs levels.
The global researches on function of the SAP and SLAM are not always identical. Some data showed that up-regulation of SLAM levels occurred on activated lymphocytes during an immune response [21-23]. It has been presumed that SLAM may play a critical role on controlling humoral immunity. Nevertheless, SLAM-deficient mice show the normal germinal center and may not be essential for T-B interaction and humoral immunity response [24]. In SAP-deficient mice model, one group observed that SAP is indispensable in CD4 T cells for germinal centers development and humoral immunity response [25]. In this current study, we observed elevated levels of both SAP and SLAM for participants who a produce efficient humoral immunity while none were changed on expression level for non-responsive subjects. It indicates that SAP may couple SLAM to protective humoral immunity response. Both of them are important for a successful humoral immune response. Based on our research result, we recognized that the modulation of SAP and SLAM expression may represent an axis by which the quality and duration of anti-HBs in humoral immunity response is regulated post immunization.
Declaration of Conflicting Interest
The authors declare that there are no conflicts of interest.
Acknowledgement
The authors would like to thank the National Natural Science Foundation of China (31500751) and the Medical Research Foundation of Zhejiang Province of China (2014KYB060) to support this study. The funder had no role in the design, analysis, or writing of this article. The Human Ethical Committee approval number in this study is ZJSJKZXLL-201401.
References
- Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 2004; 11: 97-107.
- Filippelli M. Hepatitis B vaccine by intradermal route in non-responder patients: An update. World J Gastroenterol 2014; 20: 10383-10394.
- Elefsiniotis IS, Vezali E, Kamposioras K, Pantazis KD, Tontorova R, Ketikoglou I, Moulakakis A, Saroglou G. Immunogenicity of recombinant hepatitis B vaccine in treatment-naive and treatment-experienced chronic hepatitis C patients: the effect of pegylated interferon plus ribavirin treatment. World J Gastroenterol 2006; 12: 4420-4424.
- Keeffe EB. Hepatitis A and B superimposed on chronic liver disease: vaccine-preventable diseases. Trans Am Clin Climatol Assoc 2006; 117: 227-238.
- Estevez ZC, Betancourt AA, Muzio GV, Baile NF, Silva CV, Bernal FH, Arias EP, Delhanty FA, Olazábal NM, Martín A, Batista LL, Véliz Ríos G, Hernández HH, Hernández AB, Lugo EP, de la Torre Cruz J, Batista Marchec BL, Falcón LA, Brito JT, León DO, Saura PL. Immunogenicity and safety assessment of the Cuban recombinant hepatitis B vaccine in healthy adults. Biologicals 2007; 35: 115-122.
- Wielen VM, Damme VP, Chlibek R, Smetana J, Sonnenburg VF. Hepatitis A/B vaccination of adults over 40 years old: comparison of three vaccine regimens and effect of influencing factors. Vaccine 2006; 24: 5509-5515.
- Amirzargar AA, Mohseni N, Shokrgozar MA, Arjang Z, Ahmadi N, Yousefi BM, Amanzadeh A, Shokri F. HLA-DRB1, DQA1 and DQB1 alleles and haplotypes frequencies in Iranian healthy adult responders and non-responders to recombinant hepatitis B vaccine. Iran J Immunol 2008; 5: 92-99.
- Li ZK, Nie JJ, Li J, Zhuang H. The effect of HLA on immunological response to hepatitis B vaccine in healthy people: a meta-analysis. Vaccine 2013; 31: 4355-4361.
- Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol 2006; 6: 56-66.
- Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. SAP is required for generating long-term humoral immunity. Nature 2003; 421: 282-287.
- Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL.SH2D1A regulates T-dependent humoral autoimmunity. J Exp Med 2004; 200: 261-266.
- Alem AU, Li C, Forey N, Relouzat F, Fondanèche MC, Tavtigian SV, Wang ZQ, Latour S, Yin L. Impaired Ig class switch in mice deficient for the X-linked lymphoproliferative disease gene sap. Blood 2005; 106: 2069-2075.
- Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, Cahn AP, Durham J, Heath P, Wray P, Pavitt R, Wilkinson J, Leversha M, Huckle E, Shaw-Smith CJ, Dunham A, Rhodes S, Schuster V, Porta G, Yin L, Serafini P, Sylla B, Zollo M, Franco B, Bolino A, Seri M, Lanyi A, Davis JR, Webster D, Harris A, Lenoir G, de St Basile G, Jones A, Behloradsky BH, Achatz H, Murken J, Fassler R, Sumegi J, Romeo G, Vaudin M, Ross MT, Meindl A, Bentley DR. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nature Genet 1998; 20: 129-135.
- Williams H, Macsween K, McAulay K, Higgins C, Harrison N, Swerdlow A, Britton K, Crawford D. Analysis of immune activation and clinical events in acute infectious mononucleosis. J Infect Dis 2004; 190: 63-71.
- Morra M, Barrington RA, Abadia-Molina AC, Okamoto S, Julien A, Gullo C, Kalsy A, Edwards MJ, Chen G, Spolski R, Leonard WJ, Huber BT, Borrow P, Biron CA, Satoskar AR, Carroll MC, Terhorst C. Defective B cell responses in the absence of SH2D1A. Proc Natl Acad Sci USA 2005; 102: 4819-4823.
- Cannons JL, Yu LJ, Jankovic D, Crotty S, Horai R, Kirby M, Anderson S, Cheever AW, Sher A, Schwartzberg PL. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med 2006; 203: 1551-1565.
- Veillette A, Zhang S, Shi X, Dong Z, Davidson D, Zhong MC. SAP expression in T cells, not in B cells, is required for humoral immunity. Proc Natl Acad Sci USA 2008; 105: 1273-1278.
- Cannons JL, Qi H, Lu KT, Dutta M, Gomez RJ, Cheng J, Wakeland EK, Germain RN, Schwartzberg PL. Optimal germinal center responses require a multistage T cell: B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity 2010; 32: 253-265.
- Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature 2008; 455: 764-769.
- Yin L, Alem AU, Liang J, Tong WM, Li C, Badiali M, Médard JJ, Sumegi J, Wang ZQ, Romeo G. Mice deficient in the X-linked lymphoproliferative disease gene sap exhibit increased susceptibility to murine gmmaherpesvirus-68 and hypo-gammaglobulinemia. J Med Virol 2003; 71: 446-455.
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cells require SLAM (CD150) for IL-4 production. J Immunol 2010; 185: 190-202.
- Idorenko SP, Clark EA. The dual-function CD150 receptor sub-family: the viral attraction. Nat Immunol 2003; 4: 19-24.
- Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol 2006; 6: 56-66.
- Castro AG, Hauser TM, Cocks BG, Abrams J, Zurawski S, Churakova T, Zonin F, Robinson D, Tangye SG, Aversa G, Nichols KE, de Vries JE, Lanier LL, O'Garra A. Molecular and functional characterization of mouse signaling lymphocytic activation molecule (SLAM): differential expression and responsiveness in Th1 and Th2 cells. J Immunol 1999; 163: 5860-5870.
- McCausland MM, Yusuf I, Tran H, Ono N, Yanagi Y, Crotty S. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn Kinase. J Immunol 2007; 178: 817-828.