Research Article - Biomedical Research (2017) Volume 28, Issue 10
Effect of SIRT6 on the proliferation of colorectal cancer cells
Zhibin Ma1, Yingjie Zhang2, Gang Cui2, Xiaolin Kong2, Cuicui Ren2, Shengjin Fan2, Yinghua Li2* and Jin Zhou2
1Department of Gastroenterology, the First Affiliated Hospital of Harbin Medical University, Harbin, PR China
2Department of Hematology, the First Affiliated Hospital of Harbin Medical University, Harbin, PR China
- *Corresponding Author:
- Yinghua Li
Department of Hematology
The First Affiliated Hospital of Harbin Medical University, PR China
Accepted date: March 4, 2017
Abstract
This study aimed to explore the effect of SIRT6 gene expression on the proliferation of colorectal cancer cells to improve our understanding of the pathogenesis of colorectal cancer. The colorectal cancer cell lines LOVO, SW480, SW620, LS-174T, HCT-8, and HCT-116 were employed to screen the expression of the SIRT6 gene using real-time Quantitative Polymerase Chain Reaction (Q-PCR). The expression of SIRT6 was also determined in polyp tissues, colorectal cancer tissues, and the tissues adjacent to the carcinomas using Q-PCR. After that, the SIRT6 gene was transfected into the colorectal cancer cell lines SW620 and LOVO to investigate its effects on cell proliferation, the cell cycle, and apoptosis. The expression of SIRT6 in polyp tissues was significantly higher than its expression in colorectal cancer cell lines and colorectal cancer tissue (P<0.05). However, its expression in tumors and nearby tissues showed no significant differences compared to colorectal cancer cell lines (P>0.05). There was also no significant difference between the expression of SIRT6 in the adjacent tissues and the expression in the polyp tissues (P>0.05). Following the transfection of the SIRT6 gene, the growth of both SW620 and LOVO cells was inhibited. This was due to changes in the cell cycle and significant increases in apoptosis in SW620 cells. However, no obvious apoptosis was observed in LOVO cells. Thus, the SIRT6 gene plays a role as a suppressor of the pathogenesis of colorectal cancer.
Keywords
SIRT6, Colorectal cancer
Introduction
In 2007, the third session of the Roche tumor forum reported that the incidence of colorectal cancer is increasing, and it ranked third overall among the causes of death from cancer [1]. Recent data have shown that the incidence of colorectal cancer in China ranks fourth among all malignant tumors, especially in cities with more rapid economic development. This includes Shanghai, which has ranked third overall with an annual incidence of 40 in 100,000. This is close to the incidence in developed countries in the West [2]. If colorectal cancer could be detected earlier, this would undoubtedly improve patients’ quality of life and prolong survival. Thus, exploring the pathogenesis of colorectal cancer is one of the important measures for preventing and treating colorectal cancer. The sirtuin family (SIRT1-SIRT7) extends the life of the organism mainly through the improvement of cellular physiology, including cell differentiation and survival, DNA recombination and repair, mitosis, and metabolism [3]. In cardiovascular, nervous system, and urinary system diseases, a variety of tumor-related diseases play important roles [4,5]. SIRT6 is a type of longevity factor and a member of the of the Sirtuin family. It regulates aging and other organs [6]. Furthermore, the aging process is the result of damage to cells, tissues, and organ function over time, causing organ systems to fail. A multicenter study published in Cell confirmed that the lack of SIRT6 enzyme can not only cause rapid tumorigenesis, but it also causes to abnormal glycolytic pathway function, which may also promote the formation of tumors [7,8]. One study of a liver cancer model in knockout mice confirmed the inhibitory relationship between SIRT6 and liver cancer, indicating that SIRT6 may act as a tumor suppressor and it plays a role in the gastrointestinal cancer [9-11]. Thus, we investigated SIRT6 and aimed to find a new pathogenetic mechanism underpinning colorectal cancer and its complex gene regulation network to provide insight into how to better diagnose and treat colorectal cancer.
Methods
Cell lines
The colorectal cancer cell line LOVO was obtained from the Shanghai Cell Bank of the Chinese Academy of Medical Sciences, China. SW620, SW480, LS-174T, HCT-8, and HCT-116 cell lines were from the Shanghai Life Science Research Institute of the Chinese Academy of Sciences, China. Cells were cultured in RPMI 1640 medium and maintained in an incubator with saturated humidity and 5% CO2 at 37°C. The medium was changed every 1 or 2 days. When cells entered logarithmic growth phase, they were prepared for use.
Tissue samples
Twelve patients with colorectal polyps and 12 patients with colorectal cancer treated in the Department of Digestive Endoscopy of the First Affiliated Hospital of Harbin Medical University between August 2013 and November 2013 were enrolled in this study. The colorectal polyps group included eight cases of inflammatory polyps and four adenomatous polyps. Among the 12 patients with colorectal polyps, there were seven males of an average age of 55 years and five females of an average age of 58 years. All 12 patients with colorectal cancer were pathologically diagnosed as having adenocarcinoma, including eight males of an average age of 62 years and four females of an average age of 64 years. Normal intestinal wall tissues 5 cm away from the carcinomas were sampled as “adjacent tissue” from each patient who had colorectal adenocarcinoma. All specimens were sampled, cut up immediately, and then stored at -80°C. This study was conducted in accordance with the declaration of Helsinki. This study was conducted with approval from the Ethics Committee of Harbin Medical University. Written informed consent was obtained from all participants.
Real-time PCR
Total RNA from colorectal cancer cell lines and the tissue samples was extracted using Trizol reagent according to the manufacturer’s instruction. After incubation, Trizol was added to the homogenized samples, followed by 0.2 ml of chloroform. Samples were shaken and then centrifuged at high speed for 15 min. The resulting two-layered mixture contained the upper RNA layer, and accounted for about 40% of the tube volume. The quantity of total RNA was determined using UV absorption measurements, and the purity was determined using agarose gel electrophoresis. After that, cDNAs were synthesized using the total RNA as templates for quantitative real-time PCR. The expression of SIRT6 mRNA was determined by quantitative real-time PCR and calculated using the formula RQ=2-CT. The U6 gene served as an internal control. All experiments were performed in triplicate.
Vector construction
Using the CDS sequence, specific primers for SIRT6 were designed using Primer Premier 5 software and the GeneAmp PCR System 2400 (ShangHai Ruigene Bio-pharm Technology Company). SIRT6 was amplified by PCR using the GeneAmp PCR System 2400 and separated by agarose gel electrophoresis. Target bands were recycled for double restriction enzyme digestion and ligated into a carrier vector. The vector was then transformed into competent bacteria and maintained in a 37°C incubator. Positive clones were picked for gene sequencing. After BLAST alignments, only the sequences obtained matched the original sequence with 100% identity, and were used for subsequent experiments.
Transfection
When SW620 and LOVO cells entered the logarithmic phase, the over-expression plasmid of SIRT6 was transfected using Lipo2000 kit according to the manufacturer’s instructions and incubated with 5% CO2 at 37°C. At days 0, 1, 2, 3, and 4, the proliferation rates of SW620 and LOVO cells were determined using the MTS/CCK 8 method. Apoptosis and the cell cycle dynamics of SW620 and LOVO cells were also detected using flow cytometry.
Flow cytometry
Culture medium containing SW620 or LOVO cells was placed into 15 ml conical tubes, and 2 ml PBS was used to wash the side of the tube. Next, 0.5 ml of 0.25% trypsin without EDTA was added and incubated with 0.5 ml of the pipetted cell suspension (approximately 5 × 105 cells). This was gently added to a new centrifuge tube along with 1.25 μl Annexin VFITC. This was incubated at room temperature (18-24°C) in the dark for 15 minutes. Samples were centrifuged at 1000X g at room temperature for 5 min, and then 0.5 ml pre-chilled 1X binding buffer was added to the cells to facilitate gentle resuspension. Finally, 10 μl propidium iodide was added and the samples were immediately examined for apoptosis by flow cytometry.
SW620 and LOVO colon cancer cells (1 × 106 cells each) were collected 48 h after transfection with the SIRT6 vector. Briefly, cells were washed twice with PBS after centrifugation, and pre-cooled ethanol was added to at a concentration of 70%. This was followed by fixation at 4°C overnight. The cells were then washed once with 1 ml of PBS, and then 500 μl of PI solution (50 μg/ml Propidium Bromide [PI], 100 μg/ml RNase A, 0.2% Triton X-100) was added at 4°C for 30 minutes and protected from light. This was subsequently analysed for cell cycle dynamics using flow cytometry.
Statistical analysis
The differences in the expression of SIRT6 mRNA between colorectal cancer tissues, the adjacent tissues, and the polyp tissues were analysed using a relative quantitative method. All data were processed using SPSS v19.0 statistical software. Data were shown as the means ± Standard Deviation (SD), and compared using the Wilcoxon rank test. P<0.05 was considered to be statistically significant.
Results
Expression of SIRT6 mRNA
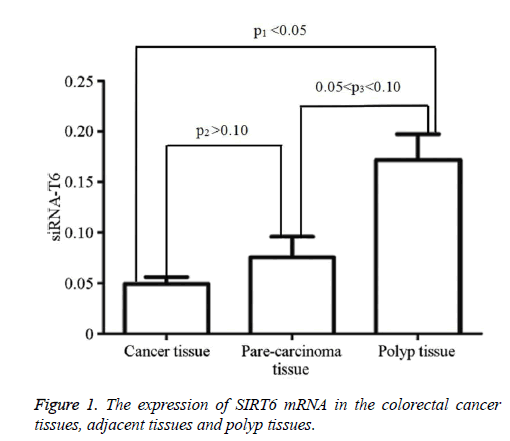
The colorectal cancer cell lines LOVO, SW620, SW480, LS-174T, HCT-8, and HCT-116 were used to determine the expression levels of SIRT6 mRNA using real-time PCR (Table 1). SIRT6 expression in the colorectal cancer tissues, adjacent tissues, and polyp tissues was also determined (Table 1). The expression of SIRT6 was highest in the polyp tissues, followed by the adjacent tissues (Figure 1).
| SIRT6 mRNA expression in colorectal cancer cell lines | SIRT6 mRNA expression in colorectal cancer tissues | SIRT6 mRNA expression in the adjacent tissues | SIRT6 mRNA expression in the polyp tissues | |
|---|---|---|---|---|
| LOVO | 1.29E-03 | 5.31E-02 | 1.80E-01 | 1.05E-01 |
| SW480 5.22E-04 | 5.22E-04 | 7.83E-02 | 1.42E-01 | 1.15E-01 |
| SW620 | 9.95E-04 | 6.33E-02 | 2.37E-02 | 1.21E-01 |
| LS-174T | 1.03E-03 | 3.81E-02 | 2.17E-01 | 1.27E-01 |
| HCT-8 | 7.96E-04 | 2.08E-02 | 1.71E-02 | 1.39E-01 |
| HCT-116 | 7.18E-04 | 2.59E-02 | 4.84E-02 | 1.51E-01 |
| 4.40E-02 | 3.36E-02 | 1.65E-01 | ||
| 6.57E-02 | 6.39E-02 | 1.70E-01 | ||
| 1.11E-02 | 1.18E-01 | 2.41E-01 | ||
| 6.26E-02 | 2.52E-02 | 3.25E-01 | ||
| 8.88E-02 | 1.33E-02 | 3.49E-01 | ||
| 4.12E-02 | 2.53E-02 | 5.27E-02 | ||
Table 1. The expression of SIRT6 mRNA in colorectal cancer cell lines and tissues.
Effect of the SIRT6 gene on cell proliferation
The over-expression plasmid for SIRT6 was transfected into the colorectal cancer cell lines SW620 and LOVO to investigate its influence on cell proliferation. After transfection, cell proliferation was assayed at five time points: days 0, 1, 2, 3, and 4. Compared to the proliferation rate of cells that did not undergo transfection, both SW620-SIRT6 and LOVO-SIRT6 cells showed lower proliferation rates at each time point (Table 2).
| Cells | Day 0 | Day 1 | Day 2 | Day 3 | Day 4 |
|---|---|---|---|---|---|
| SW620 | 100.00% | 171.23% | 245.52% | 327.37% | 447.31% |
| SW620-SIRT6 | 100.00% | 158.64% | 222.45% | 297.60% | 406.56% |
| LOVO | 100.00% | 127.43% | 159.38% | 285.38% | 336.22% |
| LOVO-SIRT6 | 100.00% | 119.05% | 138.10% | 244.18% | 275.13% |
Table 2. Cell proliferation rate of SW620 and LOVO transfected with or without SIRT6 gene.
Effect of the SIRT6 gene on inhibition rate
Compared to cells that did not undergo transfection, we found that transfection with SIRT6 had a significantly inhibitory effect on the growth of both SW620 cells and LOVO cells. This was especially true for the growth of LOVO cells, which increased in a time-dependent manner (Table 3). The inhibition rate was calculated as (1-OD of the experimental group/OD value in the control group) × 100% (the same time).
| Cells | Day 0 | Day 1 | Day 2 | Day 3 | Day 4 |
|---|---|---|---|---|---|
| SW620 | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| SW620-SIRT6 | -1.41% | 6.05% | 8.13% | 7.81% | 7.83% |
| LOVO | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% |
| LOVO-SIRT6 | 2.20% | 8.63% | 15.26% | 16.32% | 19.97% |
Table 3. Inhibition rate of SW620 and LOVO transfected with or without SIRT6 gene.
Apoptosis of colorectal cancer cell lines
We also assayed apoptosis in SW620 and LOVO cells with and without transfection of SIRT6 using flow cytometry. We found that the apoptotic index of SW620 cells increased sharply after the transfection of SIRT6, but the apoptotic index of LOVO cells increased only mildly (Table 4).
| Cells | UL | UR | LL | LR | UR+LR |
|---|---|---|---|---|---|
| LOVO | 0.58 | 2.21 | 94.78 | 2.42 | 4.63 |
| LOVO-SIRT6 | 2.72 | 2.41 | 91.5 | 2.38 | 4.79 |
| SW620 | 0.35 | 3.56 | 93.94 | 2.15 | 5.71 |
| SW620-SIRT6 | 0.51 | 6.21 | 86.3 | 6.98 | 13.19 |
Table 4. The apoptosis index of SW620 and LOVO cells.
Similar to normal cells, a bivariate flow cytometry scatter plot displays living cells in the lower left quadrant (FITC-/PI-). The upper right quadrant displays non-living cells, such as necrotic cells (FITC+/PI+). The lower right quadrant shows apoptotic cells (FITC+/PI-). With Annexin V-FITC and propidium iodide staining, living cells are not normal and they appear in the lower left corner. Early apoptotic cells only stain for Annexin V-FITC, but not propidium iodide, as shown in the bottom right corner. Late apoptotic cells and necrotic cells can be simultaneously stained by Annexin V-FITC and propidium iodide, and they are displayed in the upper right corner). The upper left corner detects errors within the allowable range. UUpper; L-Lower; L-Left; R-Right; UL: Upper Left; UR: Upper Right; LL: Lower Left; LR: Lower Right.
The cell cycles of SW620 and LOVO cells
Transfection with SIRT6 resulted in LOVO cells remaining in the G1 and S phases. In SW620 cell, it instead reduced the S and G2 phases, but did not influence the cells in the G1 phase (Table 5).
| Cells | G1 | S | G2 |
|---|---|---|---|
| LOVO | 46.85 | 49.82 | 3.33 |
| LOVO-SIRT6 | 55.81 | 44.07 | 0.12 |
| SW620 | 30.63 | 18.81 | 50.57 |
| SW620-SIRT6 | 37.91 | 14.03 | 48.06 |
Table 5. Cell cycles of SW620 and LOVO cells with and without transfection with SIRT6 gene.
Discussion
SIRT6 is a member of the Sirtuin family, which is a class of proteins that are highly evolutionarily conserved from simple bacteria to eukaryotes. They are considered to be “longevity genes” [9]. In higher animals, the Sirtuin family includes seven members: SIRT1-SIRT7 [10,11]. Among these, SIRT1, -6, and -7 are mainly expressed in the nuclei. SIRT2 is expressed in both the nuclei and the cytoplasm. SIRT3, -4, and -5 are mainly expressed in the mitochondria [12]. The human SIRT6 gene is located on chromosome 19 (19pl3.3), and it expresses and transmits its functions by encoding the protein. The molecular weight of the SIRT6-encoded protein is 39.1 kDa, and it has a pI of 9.12 [13]. However, in eukaryotes, the cytoplasm also contains a small amount of genetic material from the chromosomes, which can participate in nuclear functions [14]. As a result, SIRT6 is also expressed at low levels in the cytoplasm.
SIRT6 is a chromosome-associated protein necessary for Basic Excision Repair (BER) of damaged DNA and the maintenance of gene stability [15]. Thus, it plays important roles in DNA damage and repair, telomerase function, and cellular metabolic processes [16-18]. The loss of SIRT6 causes the excision of gene segments and other chromosomal aberrations [19], as well as an increase in the susceptibility to pathogenic and carcinogenic factors. Studies have found that SIRT6 can participate in the regulation of glucose metabolism in vivo. It can interfere with glucose absorption and glycolysis by inhibiting growth factors and Hif1 [20,21].
The expression of SIRT6 in colorectal cancer has seldom been reported. Currently, studies of SIRT6 mainly focus on aging and genital gland embryonic tumors. It has been shown that SIRT6 also has NAD+-dependent deacetylase activity and can regulate the stability of chromosomes [22-24]. It substrate is H3K9 and H3K56. In rats, the lack of SIRT6 can shorten life and cause premature aging symptoms [25]. In humans, mutations in SIRT6 and uncoupling protein 5 are associated with the formation of carotid artery atherosclerosis plaques. SIRT6 can positively regulate the expression of tumor necrosis factors after transcription, and it participates in acute or chronic inflammation [26].
In 2010, it was reported that tumor cells grow mainly using energy obtained from glycolytic pathways [27]. Researchers suggested that SIRT6 is a key factor that activates the glycolytic pathway. Thus, we speculated that ablation of SIRT6 will disrupt the glycolytic pathway and, thus, help control tumor growth.
We examined the expression of SIRT6 mRNA in colorectal cancer cell lines, colorectal cancer tissues, adjacent tissues, and polyp tissues. The results showed that SIRT6 mRNA is lowly expressed in colorectal cancer cell lines and colorectal cancer tissues. It is significantly lower than the expression observed in the polyp tissues (P<0.05, Table 1 and Figure 1), indicating that the SIRT6 gene plays a role as a tumor suppressor in colorectal cancer. However, the expression of SIRT6 in polyp tissues and tissues adjacent to carcinomas showed no statistical significance (P>0.05). The reasons for this may be that the tissues adjacent to the carcinoma may also increase their overall energy intake and activate the glycolytic pathway. It is also possible that the benign polyp tissues have a higher growth rate than normal intestinal mucosa, which requires more energy. To explore the significance of the differential expression of SIRT6 in the above-mentioned tissues, the sample size should be expanded and the adjacent tissues should be sampled further away from the tumor.
In this study, we successfully established SW620-SIRT6 and LOVO-SIRT6 cell models. After 4 days of culture, we found that the proliferation rates for both SW620-SIRT6 and LOVOSIRT6 cells significantly decreased when compared with cells that did not undergo transfection. The proliferation rate of LOVO-SIRT6 cells was 406% higher. However, this was significantly lower than that of LOVO cells (447%, P<0.05). Meanwhile, the proliferation rate of SW620-SIRT6 cells was 275%, which was significantly lower than that of SW620 cells (336%, P<0.05). These results illustrated that the SIRT6 gene can inhibit the proliferation of colorectal cancer cells. Next, we analysed the inhibition rates following transfection with the SIRT6 gene, and we found that 19.97% of LOVO cells and 7.83% of SW620 cells experienced proliferation inhibition after transfection with SIRT6. The amount of inhibited cells increased markedly upon extension of the incubation time, indicating that SIRT6 inhibits the proliferation of colorectal cancer cell lines in a time-dependent manner, particularly for the LOVO cell line. However, based upon the cell apoptosis index, transfection with the SIRT6 gene increased apoptosis in LOVO cells from 4.63 to 4.79 and it increased it in SW620 cells from 5.71 to 13.19. This indicated that SIRT6 induced more apoptosis in SW620 cells than in LOVO cells. In addition, the cell proliferation curve produced by flow cytometry also showed that the transfection of SIRT6 mainly inhibited the growth of LOVO cells at the G1 and S phases, while it inhibited the growth of SW620 cells at the S and G2 phases. These are both stages for the synthesis and replication of DNA, suggesting that SIRT6 has different inhibitory roles in the various colorectal cancer cell lines (Tables 2 and 3). Nevertheless, the decreased proliferation observed in SW620- SIRT6 and LOVO-SIRT6 cells indicated that SIRT6 inhibits the growth of colorectal cancer by promoting apoptosis in colorectal cancer cells. However, for the different colorectal cancer cell lines, SIRT6 exerted its inhibitory effects in different ways. It inhibited the growth of LOVO cells at the G1 and S phases, but not at the G2 phase. Meanwhile, it exerted different inhibitory effects on the SW620 cell line, which displayed a decreased number of cells at each phase of the cell cycle, but this was not as significant as LOVO cells. This suggested that the tumor suppressor functions of SIRT6 in the different phenotypes observed in colorectal cancer cell lines are also different. The underlying mechanisms need to be confirmed by more experimental studies (Tables 4 and 5).
Overexpression of SIRT6 can cause apoptosis in a wide variety of tumor cells. It inhibits the glycolysis pathway and interrupts the energy supply chain for tumor cells. Thus, tumor cells become apoptotic due to the lack of energy reserves. However, in this study, we found that SIRT6 plays different inhibitory roles in different kinds of colorectal cancer cells, suggesting that its effects are multifaceted and its regulatory mechanism is complex. Studies have reported that a reduction in energy intake, such as a 25% reduction in the total calorie consumption, can reduce the expression of SIRT6. SIRT6 enzyme can also promote the secretion of Tumor Necrosis Factor alpha (TNF-α) and enhance apoptosis in tumor cells. SIRT6 is not only an important regulator of mammalian lifespan and a known modulator of cellular glucose metabolism. It is also a very important tumor suppressor. These findings may help develop treatments that directly target cell metabolism, which may help to suppress the growth of tumors. In fact, SIRT6 inhibits the growth of tumor cells during antiaging processes, regulates metabolism, and promotes apoptosis. There is a clear relationship amongst these functions.
Among gastrointestinal tumors, the incidence of colorectal cancer is increasing gradually and it is gaining greater attention. It has been confirmed that colorectal cancer cells use the Warburg effect [28,29] to obtain energy through glycolysis. This is similar to other tumors. Based upon our analysis, SIRT6 may function as a regulatory switch for glycolysis. It may suppress the growth of tumors by controlling glycolysis. Thus, we suggest that SIRT6 may be a tumor suppressor gene. Our experimental results aimed to identify a molecular mechanism underpinning the development of colorectal cancer. These results may help identify a drug that targets this mechanism. This work provides a solid foundation for exploring the incidence theory and improving the clinical care of colorectal cancer.
Conflict of Interest
All authors have no conflict of interest regarding this paper.
References
- Liu M, Shen S. Colorectal carcinoma and DNA methylation. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2009; 34: 1266-1270.
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014; 64: 104-117.
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J 2007; 404: 1-13.
- Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, Gao B, Deng CX. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab 2010; 12: 224-236.
- Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell 2010; 9: 162-173.
- Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 by enforced expression of micro-RNA miR-125a or miR125-b. J Biol Chem 2007; 282: 1479-1486.
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006; 124: 315-329.
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012; 483: 218-221.
- Andre T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan P, Bridgewater J, Tabah-Fisch I, de Gramont A. Multicenter International Study of Oxaliplatin/5-Fluorouracil/Leucovorin in the Adjuvant Treatment of Colon Cancer (MOSAIC) Investigators. Oxaliplatin, fluoroura-cil and leucovorinas adjuvant treatment for colon cancer. N Engl J Med 2004; 350: 2343-2351.
- Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene 2006; 25: 4633-4646.
- Harder J, Engelstaedter V, Usadel H, Lassmann S, Werner M, Baier P, Otto F, Varbanova M, Schaeffner E, Olschewski M, Blum HE, Opitz OG. CpG-island methylation of the ER promoter in colorectal cancer: analysis of micrometastases in lymph nodes from UICC stage I and II patients. Br J Cancer 2009; 100: 360-365.
- Kelly G. A review of the sirtuin system, its clinical implications, and the potential role of dietary activators like resveratrol: part 1. Altern Med Rev 2010; 15: 245-263.
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 2000; 273: 793-798.
- Hall JA, Dominy JE, Lee Y, Puigserver P. The sirtuin familys role in aging and age-associated pathologies. J Clin Invest 2013; 123: 973-979.
- Haigis MC, Guarente LP. Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev 2006; 20: 2913-2921.
- Mahlknecht U, Ho AD, Voelter-Mahlknecht S. Chromosomal organization and fluorescence in situ hybridization of the human Sirtuin 6 gene. Int J Oncol 2006; 28: 447-456.
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 2005; 16: 4623-4635.
- Yu SS, Cai Y, Ye JT, Pi RB, Chen SR, Liu PQ, Shen XY, Ji Y. Sirtuin 6 protects cardiomyocytes from hypertrophy in vitro viainhibition of NF-?B-dependent transcriptional activity. Br J Pharmacol 2013; 168: 117-128.
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008; 452: 492-496.
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009; 8: 2664-2666.
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 2009; 8: 2662-2663.
- Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med 2008; 263: 128-141.
- Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, Jou W, Gius D, Deng CX. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem 2010; 285: 36776-36784.
- Zhong L, DUrso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 2010; 140: 280-293.
- Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle 2011; 10: 3153-3158.
- Dong C, Della-Morte D, Wang L, Cabral D, Beecham A. Association of the sirtuin and mitochondrial uncoupling protein genes with carotid plaque. PLoS One 2011; 6: 27157.
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012; 21: 297-308.
- Van Gool F, Gallí M, Gueydan C, Kruys V, Prevot PP, Bedalov A, Mostoslavsky R, Alt FW, De Smedt T, Leo O. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat Med 2009; 15: 206-210.
- Chen Z, Lu W, Garcia-Prieto C, Huang P. The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr 2007; 39: 267-274.