- Biomedical Research (2015) Volume 26, Issue 4
Effect of Mutation S34N on hCAP in Periodontal Dental Arthritis Patients: Molecular Dynamics Simulation Study
Wei-Qun Zhang1*, Yun-Qi Liu2,Quan-Sheng Ma1, Li Wang1, Zhao-Hui Hu1
1Department of Oral Implantology, Provincial Hospital Affiliated to Shandong University, Jinan, Shandong province, 250021, China
2Department of Nephrology, hospital affiliated to Binzhou Medical University, Binzhou 256600, China
- *Corresponding Author:
- Wei-Qun Zhang
Department of Oral Implantology
Provincial Hospital Affiliated to Shandong University
No. 324 of Jingwei Road, Huaiyin District
Jinan, Shandong 250021, China
Tel: 0086-531-68776585
Fax: 0086-531-68776585
E-mail: zwqzhangweiqun@hotmail.com
Abstract
Studies have shown that genetic factors involved in the host responses might determine the
severity of periodontitis and the missense mutation of S34N in human cathelicidin antimicrobial
peptide (hCAP) is one of the predominant factors in this severity. The aim of this study is to
study the effect of this mutation on protein dynamics. The FASTA sequence of the protein was
subjected to four different (SIFT, Polyphen-2, PhD-SNP and MutPred ) polymorphism effect
prediction servers and the same format of the sequence was used to modelled the three
dimensional structure of hCAP. The modelled structure was then subjected to Molecular
dynamics simulations for Atomic insight. All the four servers predicted the mutation to be
damaging and on analysing the effect of this mutation with the help of GROMACS, the results
were showing the mutant structure unstable and differencing at every level in comparison with
wild type hCAP.
Keywords
Periodontitis; hCAP; Mutation; Molecular Dynamics Simulation
Introduction
Periodontal diseases are characterized by localized infections and inflammatory conditions that directly affect teeth supporting structures which are the major cause of tooth loss [1]. Several studies have demonstrated the involvement of autoimmune responses in periodontal disease [2-7]. Bacteria in the dental plaque induce antibody formation, Autoreactive T cells, natural killer cells; ANCA, heat shock proteins, autoantibody, and genetic factors are reported to have an important role in the autoimmune component of periodontal disease [8]. Studies have suggested that genetic factors are important determinants of susceptibility to periodontitis since they determine and modify host responses to the microbial challenge [9, 10]. Any anomaly in the immune responses against bacterial stimulus may cause variations in the susceptibility for periodontitis [11]. The severity of periodontitis is dependent on genetic factors involved in host responses [12, 13].
Microbial dental plaque is the major factor causing periodontitis and many studies have shown that genetic factors play a crucial role in the susceptibility for aggressive periodontitis [14]. hCAP (Human cathelicidin antimicrobial peptide) an 18kDa protein is the only known cathelicidin for humans, which act as an antimicrobial agent [15,16] by binding to the microbial cell membrane and killing the bacteria by disrupting its metabolism[17]. This protein is an important defence molecule in the oral cavity as its epithelia is at constant expose to microbial pathogens [18] and its presence in human tongue, buccal mucosa and saliva also underlines its importance in oral immunity [19,20]. The hCAP is expressed by CAMP gene, which has been found to carry missense mutation at codon 34 i.e. p.S34N which seems to be contributing factor for generalized aggressive periodontitis [21].
Our aim here in this study is to explore the effect of this mutation on protein dynamics using molecular dynamics simulation simulations (MDS) tool. THE tools has been used to understand the effect of the mutation on the protein at atomic level [22,23].Various polymorphism effect prediction servers available online: SIFT [24], Polyphen-2 [25], PhD-SNP [26] and MutPred [27] were also used to elucidate the effect.
Material and Method
Protein Modelling
As the crystal structure of hCAP is not available, Molecular modelling using I-TASSER [28] an online protein modelling server was used. The FASTA sequence of the 170 amino acids was subjected to the server and the model with the best C-score was selected for further study.
Polymorphism damage prediction
Four in silico tools were selected meticulously so as each factor is looked into and double checked by other tool which uses different algorithm to predict the effect of the polymorphism on protein structure and function.
Molecular Dynamics Simulation
The MDS studies were performed by Gromacs 4.5.3 package [29]. For Wild type (wt) hCAP the modelled structure was used as a starting structure for MDS and the Accelrys Discovery Studio [30] was used to make single point mutation at codon 34 to get mutant (mu) structure. The structures obtained were then subjected to GROMOS96 43a1 force field and then placed in a model of a pre-equilibrated water bath. To obtain the ionic neutral environment, counter-ions were added using the “genion” tool of gromacs package. Both the ions and solvent molecules of both the wt and mutant system were restrained to the original position with a force constrain of 100Kcal/mol for 5000 steps before being subjected to energy minimization for 5000 iteration. Regular Berendsen temperature coupling method [31] was used for regulating the temperature inside both the systems. Electrostatic interactions were computed using the Particle Mesh Ewald method [32]. The non-bonded pair list was updated every five steps and conformations were stored every two pico seconds (ps). Position restraint simulation for 1000 ps was implemented to allow solvent molecules to enter the cavity region of structure. Finally, system was subjected to MDS for twenty nano seconds (ns). Root mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of gyration (Rg) and Principal component analysis (PCA) were carried out by using inbuilt gromacs tools. g-hbond was used to calculate number of distinct hydrogen bonds formed by specific residues to other amino acids within the protein during the simulation (NH bond). g_sham was used extensively used to obtain free energy landscape. The graphs were plotted using Grace GUI toolkit 5.1.22 version. The free energy land scapes were plotted using gnuplot 4.6.0 version. All the visualizations were carried out using Pymol, Ligplus, VMD [33] and graphs were plotted using Grace Program [34] and GNUPlot. The trajectories were analyzed using the inbuilt tool in the GROMACS distribution.
Result and Discussion
The protein structure was modelled using I-TASSER, an online server which uses threading method. The best model based on C-score had good quality and resolution (Figure 1) was selected and the subjected to PROCHECK in SAVES server to check its psi-phi distribution, the amino acids arranged in most favoured regions in plot were 89.2% and some of the amino acids in disallowed regions of 2.3%.
Disease association with amino acid polymorphism is difficult, as the polymorphism can have a vast number of effects on the gene product. These effects can be in stability, structure, disruption in catalytic active site, ubiquitination site etc. The Four Insilico tools that we used for this study gave us an idea about the possibility of the disease association of the S34N polymorphism. The details of the servers that are used in our study and the results generated are described in where there algorithm, working and criteria for selection is give. All the servers showed the polymorphism to be damaging.
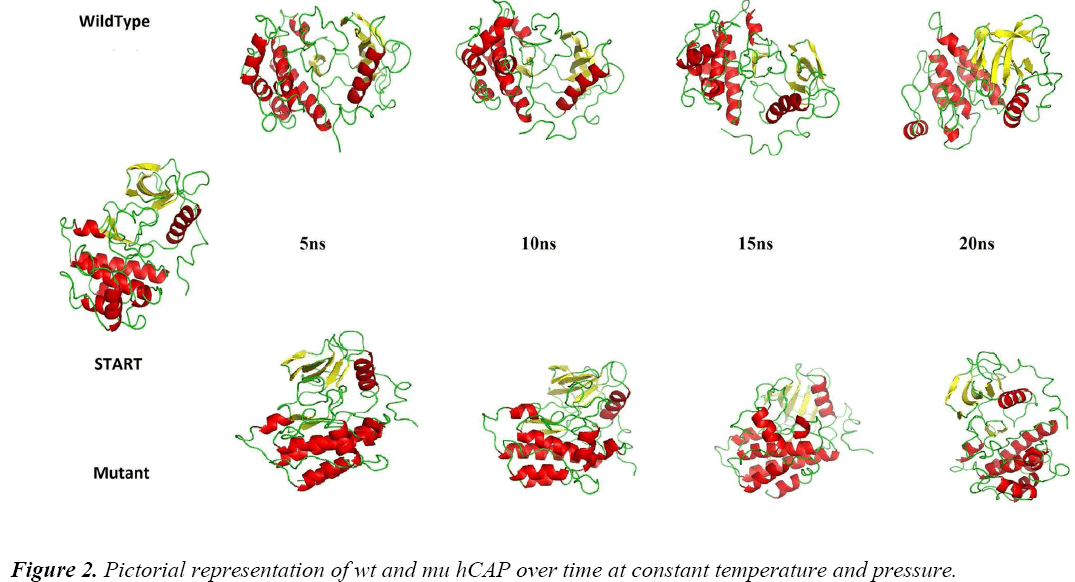
To analyse the effect of the polymorphism of S34N at atomic level, MDS approach was used. Figure 2 shows the difference between the two structures at different intervals of time at constant temperature and pressure during the simulations, showing the change in protein structure due to mutation. For further analysis Gromacs inbuilt tools g_rmsf, g_rms, g_ gyrate, g-covar and g_anaeig were used.
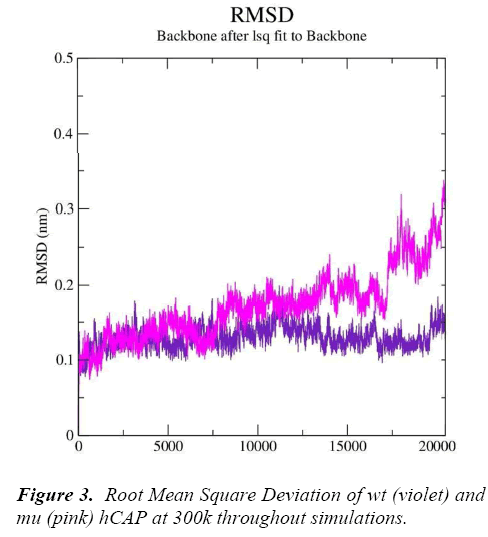
g_rms gave us the RMSD (root mean square deviation) value curves of wt and mu hCAP during the simulations, Figure 3 represents RMSD fluctuations of both wt and mu structures over twenty thousand picoseconds. The analysis is indicating that mutation at codon 34 from S to N is effecting the conformation of the hCAP, with mutant structure showing more deviation pattern than native in general.
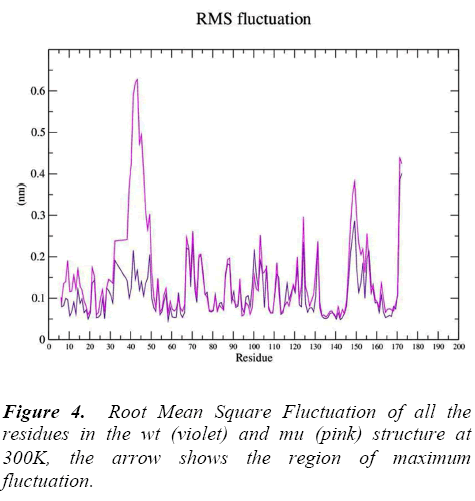
g_rmsf was used to calculate the atomic standard deviation .From this analysis it is evident that mutation at codon 34 from S to N is making the mutant structure more fluctuating (Figure 4) particularly at residue 34 , 50, 140 and 160 .These fluctuations in the mu structure is over all and not specified to the region of polymorphism.
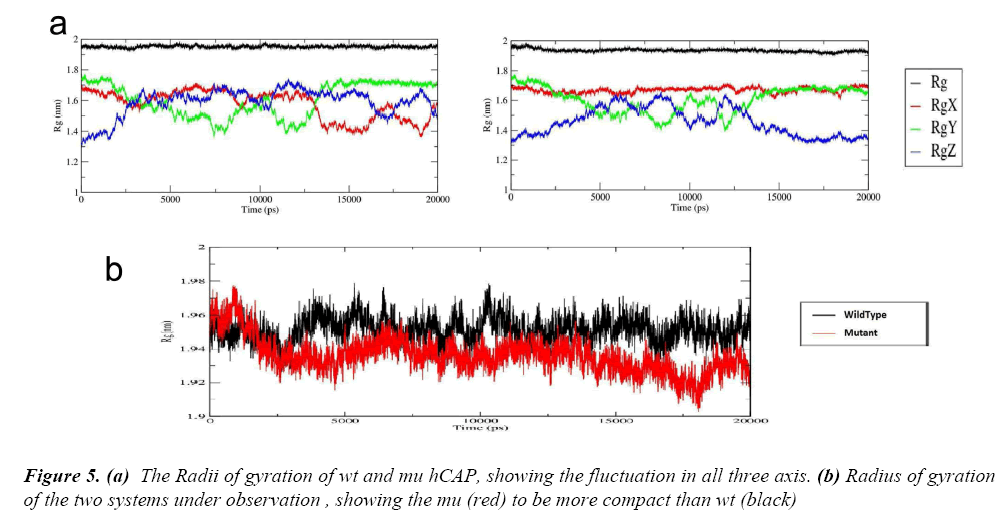
After these two interlinked analysis, change in radius of gyration of the two structures was calculated using g_gyrate tool. The tool analyzes the shape of the protein over time at constant temperature and pressure, the results generated show that the mutant structure is more compact than the native hCAP .In Figure 5(a) the black represents the native structure and red the mutant, Figure 5(b) and 5(c) are the representation of Radii of gyration of native and mutant structure over the three axes.
To further understand the effect of the S34N mutation on hCAP, g_hband tool was used. The intra-protein hydrogen bond pattern over time was studies. The native protein is forming about 120 out of 52344 possible average number of intra-protein hydrogen bonds per timeframe, while as mutant is forming 130 out of 52344 possible, The result is in accordance with Rg of both structures over time, the increase in number of hydrogen bond in corroborating with the mutant structure being more compact.
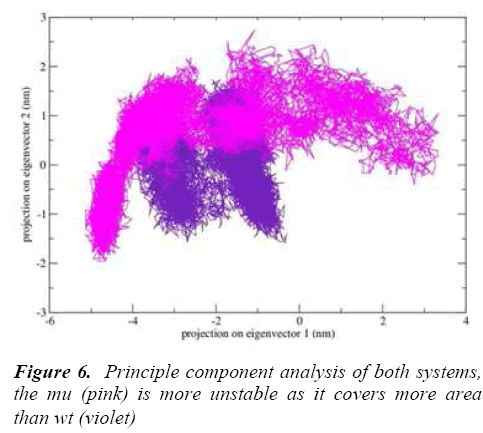
g-covar and g_anaeig tools were used for understanding the effect of this mutation on global correlated motions in atomic simulations. The figure 6 depicts the projection of principal component 1 vs. principle component 2 of both structures, the cluster obtained from wt structure is stable , where as the projection of first two PC of both mutants is covers a large area.
Conclusion
The authors would like to thank the management of Shandong University for their support in the study. Further we like to acknowledge the bioinformatics division of Binzhou Medical University for their support in this study.
Acknowledgement
The amino acid variation from S to N at codon 34 in hCAM is damaging the native conformation. The change in RMSD and Rg is conclusively defining the change in conformation of hCAM mutant structure. The change in RMSF in mutant structure is also confirming the variation in structural dynamics. These changes can be pivotal in varying the anti microbial activity, which is ultimately leading to aggressive periodontitis associated phenotypes.
References
- Newman MG, Takei H, Klokkevold PR, and Carranza FA. Carranza's clinical periodontology. Elsevier health sciences (2011)
- Jiang S and Lechler RI. Regulatory T cells in the control of transplantation tolerance and autoimmunity. Am J Transplant 2003; 3(5): 516-524.
- Anusaksathien O and Dolby AE. Autoimmunity in periodontal disease. J Oral Pathol Med 1991; 20(3): 101-107.
- Anusaksathien O, Singh G, Matthews N, and Dolby AE. Autoimmunity to collagen in adult periodontal disease: immunoglobulin classes in sera and tissue. J Periodontal Res 1992; 27(1): 55-61.
- Hahn CL, Schenkein HA, and Tew JG, Polyclonal B. Cell acivators and in vitro induction of auto antibody reactive with collagen. J Periodontal Res 1991; 32: 608-613.
- Wucherpfennig KW. Mechanisms for the induction of autoimmunity by infectious agents. J Clin Invest 2001; 108(8) : 1097-1104.
- Reinholdt J, Bay I, and Svejgaard A. Role of HLA antigen in periodontal disease, J Dent Res 1977; 56(10): 1261-1263.
- Encinas JA and Kuchroo VK. Mapping and identification of autoimmunity genes, Curr Opin Immunol 2000; 12(6): 691-697.
- Nair S, Faizuddin M, and Dharmapalan J. Role of Autoimmune Responses in Periodontal Disease. Autoimmune Dis 2014; 1-7.
- Page RC, Offenbacher S, Schroeder HE, Seymour GJ, Kornman KS. Advances in the pathogenesis of periodontitis: summary of developments, clinical implications and future directions. Periodontol 2000 1997; 14: 216-248.
- Michalowicz BS, Wolff LF, Klump DG, Hinrichs JE, Aeppli D, Bouchard TJ, et al. Periodontal findings in adult twins. J Periodontol 1991; 62(5): 293–9.
- Gore EA, Sanders JJ, Pandey JP, Palesch Y, Galbraith GM. Interleukin-1beta13953 allele 2: association with disease status in adult periodontitis. J Clin Periodontol 1998; 25(10): 781-785.
- Laine ML, Farre MA, Gonzalez G, van Dijk LJ, HamAJ, Winkel EG, et al., Polymorphisms of the interleukin-1 gene family, oral microbial pathogens, and smoking in adult periodontitis. J Dent Res 2001; 80(8): 1695-1699.
- Loos BG, Van der Velden U, Laine ML. Susceptibility. In: Lindhe J, Karring T, Lang NP, editors. Clinical periodontology and implant dentistry. 5th ed. Oxford: Blackwell Munksgaard 2008; pp328– 46.
- Durr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta 2006; 1758(6): 1408-1425.
- Bals R, Wilson JM. Cathelicidins – a family of multifunctional antimicrobial peptides. Cell Mol Life Sci 2000; 60(4): 711-720.
- Ciornei CD, Sigurdardo´T, Schmidtchen A, Bodelsson M. Antimicrobial and chemoattractant activity, lipopolysaccharide neutralization, cytotoxicity, and inhibition by serum of analogs of human cathelicidin LL-37. Antimicrob Agents Chemother 2005; 49(7): 2845–50.
- Dale BA, Kimball JR, Krisanaprakornkit S, Roberts F, Robinovitch M, O’Neal R, et al. Localized antimicrobial peptide expression in human gingival. J Periodontal Res 2001; 36(5): 285–94.
- Frohm NM, Sandstedt B, Sorensen O, Weber G, Borregaard N, Stahle-Backdahl M. The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6, Infect Immun 1999; 67(5):2561–6.
- Murakami M, Ohtake T, Dorschner RA, Gallo RL. Cathelicidin antimicrobial peptides are expressed in salivary glands and saliva. J Dent Res 2002; 81(12): 845–50.
- Türkoglu O, Berdeli A, Emingil G, Atilla G, A novel p. S34N mutation of CAMP gene in patients with periodontal disease. Arch oral boil2011; 56(6); 573- 579.
- Chikan NA, Bukhari S, Shabir N, Amin A, Shafi S, Qadri RA, & Patel TN. Atomic Insight into the Altered O6-Methylguanine-DNA Methyltransferase Protein Architecture in Gastric Cancer. PloS one 10.5 (2014): e0127741-e0127741.
- Bukhari S, Mokhdomi TA, Chikan NA., Amin A, Qazi H, Wani SH, et al. Affinity proteomics led identification of vimentin as a potential biomarker in colon cancers: insights from serological screening and computational modelling. Molecular BioSystems 2015; 11(1): 159-169.
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4(7):1073- 81.
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Met 2010; 7(4): 248-249.
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006; 22: 2729-2734.
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009; 25: 2744–2750.
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008; 9: 40.
- Hess B, Kutzner C, Van Der Spoel D, & Lindahl E, GROMACS 4: Algorithms for highly efficient, loadbalanced, and scalable molecular simulation. J chem theory comput 2008; 4(3): 435-447.
- Accelrys Software Inc., Discovery Studio Modeling Environment, Release 3.5, San Diego: Accelrys Software Inc., 2012.
- Berendsen HJC, Postma JPM, van Gunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phy 1984; 81: 3684–3690.
- Cheatham TE, Miller JL, Fox T, Darden TA, Kollman PA. Molecular dynamics simulations on solvated biomolecular systems: the particle mesh ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc 1995; 14: 4193–4194.
- Humphrey W, Dalke A, & Schulten K. VMD: visual molecular dynamics. J mol graph 1996; 14(1): 33-38.
- Turner PJ. XMGRACE, Version 5.1. 19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR. (2005)