- Biomedical Research (2014) Volume 25, Issue 2
Distribution of the multidrug efflux pump genes adeA, adeI, adeJ, adeY and integrons in clinical isolates of Acinetobacter baumannii from Malaysian hospitals.
Sue-Bee Kor, Beatrice-Sing-Yieng Tou, Cornelius-Kwang-Lee Chieng, Michele-Sook-Yuin Hiew and Choy-Hoong Chew*Department of Biomedical Science, Faculty of Science, Universiti Tunku Abdul Rahman, Jalan Universiti, Bandar Barat, 31900 Kampar, Perak, Malaysia
- *Corresponding Author:
- Choy-Hoong Chew
Department of Biomedical Science
Faculty of Science, Universiti Tunku Abdul Rahman
Jalan Universiti, Bandar Barat
31900 Kampar, Perak, Malaysia.
Accepted date: November 20 2013
Abstract
The objective of this study was to evaluate the incidence of multidrug resistance nodulationcell division pump (RND pump) genes and integrons in multidrug-resistant Acinetobacter baumannii isolated from in-patients in Malaysian hospitals. A total of 43 clinical isolates of A. baumannii were examined. All samples were found to be 100% resistant to ampicilin (n=43), ceftriaxone (n=43) and cefuroxime (n=43), followed by augmentin (n=40) and chloramphenicol (n=40) (93%). Local isolates of A. baumannii were also found to develop resistance towards the new class of antibiotic-tigecycline (58.14%). The presence of adeA, adeIJ, adeY RND efflux system genes and integrase gene was assessed by polymerase chain reaction (PCR). adeY was not detected in any of the samples. However, majority of the isolates (62.7%, n=27) carried adeA and adeIJ genes, but not integrase gene. Interestingly, the isolates harboring these genes also showed the highest level of resistance toward all antibiotics. In addition, 13.9% of the isolates (n=6) did not carry any of the RND genes; while class 1 integrons were only detected in three isolates (7%, n=3), in which all harbored the same gene cassette aacA4-catB8-aadA1. This suggests the possibility of higher usage of RND efflux pump genes in antibiotic resistance mechanism for local A. baumannii compared to integron genes. Besides, the co-presence of adeA and adeIJ genes also reveals their possible relationship for antimicrobial resistance in A. baumannii isolates.
Keywords
Acinetobacter baumanii, RND efflux pump genes, Integrons, Multidrug resistance
Introduction
Acinetobacter baumannii, a Gram-negative opportunistic pathogen, is the agent that frequently causes various types nosocomial infections, including urinary tract infections, meningitis and bacteremia in hospitalised patients [1-3]. It has been observed to be resistant to all existing classes of antibiotics [2], including glycylcyclines; a new class of modified tetracycline antibiotics [4].
Resistance-nodulation division (RND) family, a member of active efflux pump mechanism has been known to play an important role in multidrug resistance in Gramnegative bacteria. The efflux transporters are organized as multicomponent system where it is protected by an outer membrane and the efflux pump works in conjunction with a periplasmic fusion protein together with an outer membrane protein [5]. To date, five kinds of RND system has been described in Acinetobacter sp. [6-9]. Indeed, Ade-ABC efflux pump is well characterized. The increased expression of the adeABC genes has led to resistance to many antibiotics including aminoglycosides, betalactams, chloramphenicol, erythromycin, tetracyclines, and the dye ethidium bromide [6-10]. However, studies on the other two RND pump genes, adeIJK and adeXYZ are limited as compared to adeABC. Structural genes for adeIJK have been perceived to contribute to intrinsic lowlevel of resistance to several antibiotics in A. baumannii [11]. The role of AdeXYZ in antimicrobial resistance, on the other hand, has yet to be ascertained.
Integrons, first described by Stokes and Hall [12], have strong linkage with multidrug resistance in Gramnegative bacteria [13]. The association of the integrondriven capture system with mobile elements such as transposons and plasmids has facilitated the spread of antibiotic resistance genes between species significantly. To date, five distinct classes of integrons have been described [14]. Class 1 integrons are commonly found in clinical isolates of Gram-negative bacilli, including A. baumannii [15,16]. In this study, we assessed the distribution of RND pump genes adeA, adeI, adeJ and adeY, and integrase genes in A. baumannii collected from hospitals around peninsular Malaysia, and analyzed their antibiotic susceptibility pattern. Such typing can be used to further characterize isolates and should prove to be very useful in epidemiological studies.
Materials and Methods
Bacteria strains and growth media
A total of 43 multidrug resistant (MDR) clinical A. baumannii strains were collected from various hospitals around Malaysia. These isolates were obtained from wound (n=10), pus (n=4), sputum (n=8), blood (n=1), swab (n=9) and others (n=11). The isolates were routinely grown at 37ºC on Luria Bertani (LB) agar.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined according to the Clinical and Laboratory Standards Institute (CLSI) using the Kirby-Bauer disk diffusion assay on Mueller Hinton agar (Oxoid, UK) in accordance to the described method in Cheong et al. [17]. The susceptibility profile was determined for 15 antibiotics: Amikacin (30 μg), Amoxicllin/ Clavulanic acid (Augmentin, 3 μg), Ampicillin (10 μg), Ceftazidime (30 μg), Ceftriaxone (30 μg), Cefuroxime (30 μg), Chloramphenicol (30 μg), Ciprofloxacin (5 μg), Gentamicin (10 μg), Imipenem (10 μg), Meropenem (10 μg), Norfloxacin (10 μg), Tetracycline (30 μg), Tigecycline (15 μg), and Trimethoprim/Sulfamethoxazole (Co-trimoxazole, 25 μg). All antibiotic discs were purchased from Oxoid, United Kingdom.
Preparation of DNA template
Total DNA template was prepared using fast-boiling method. Overnight bacterial culture was centrifuged at 12,000 rpm for 5 min and pellet was re-suspended with 300 μl of sterile deionized water. The bacterial suspension was then boiled for 5 min and placed immediately on ice for another 2 min before before centrifuging at 12,000 rpm for 2 min. The supernatant was aspirated and used for subsequent polymerase chain reaction.
Amplification of efflux pump genes
The presence of adeA, adeI, adeJ and adeY genes of RND efflux pump system was determined by PCR. The primers used and their references are listed in Table 1. PCR reaction was performed in a final volume of 25 μl containing PCR buffer, 2 mM of MgCl2, 0.4 μM of each primers, 0.4 mM of dNTP mix, 1 U of Taq polymerase (Promega, USA) and 250 ng/μl of template DNA. PCR reaction was performed in thermal cycler (Biometra, Germany) with the parameter: initial denaturation 94ºC for 5 min, 30 cycles of 94ºC for 60 sec, 56ºC for 60 sec and 72ºC for 60 sec, and a final elongation at 72ºC for 7 min. The presence and sizes of the amplicons were assessed by electrophoresis in 1.5% agarose gels stained with ethidium bromide.
Integrons, gene cassettes amplification and restriction fragment length polymorphism
Integrons were detected using degenerate primers, hep35 and hep36, which hybridize to the conserved region of integrase genes intI1, intI2, and intI3. Class of integrons was further determined by performing restriction fragment length polymorphism (RFLP) using HinfI and RsaI [18,19]. To detect for the inserted gene cassettes, 5’CS and 3’CS primers were used to amplify the variable regions of class 1 integrons [19,20]. The amplification protocol used was: denaturation step at 94ºC for 5 min, 30 cycles of 94ºC for 30 sec, 55ºC for 30 sec and 72ºC for 45 sec for integrase gene or 90 sec for amplification of gene cassette.
Results
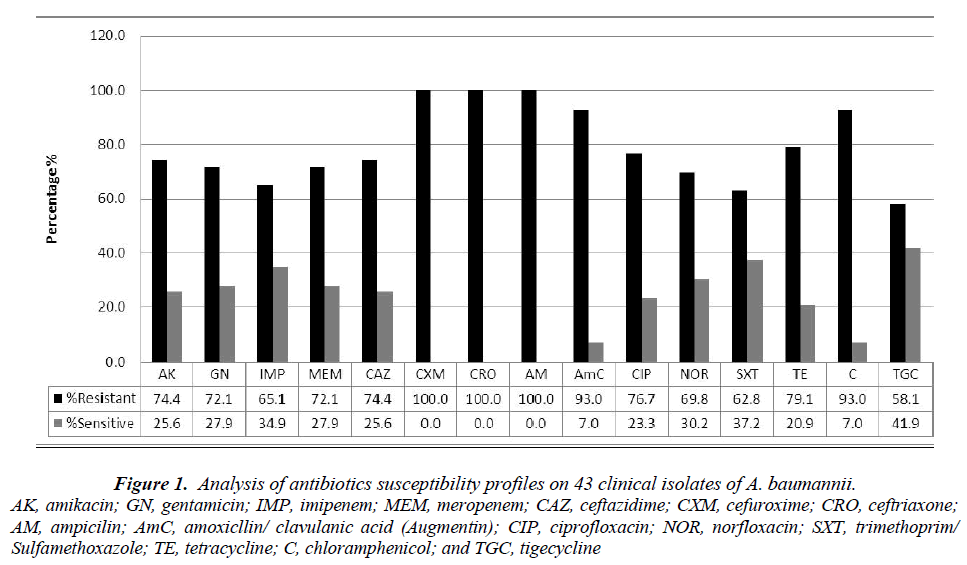
Figure 1 shows the antimicrobial susceptibility profiles of the 43 clinical isolates of A. baumannii. As observed, all the isolates (100%) were resistant to ampicillin, ceftriaxone and cefuroxime. This was followed closely by augmentin and chloramphenicol (93%), tetracycline (79.1%), amikacin and ceftazidime (74.4%), and gentamicin and meropenem (72.1%). More than 58.1% of the isolates showed resistance to tigecycline.
Figure 1: Analysis of antibiotics susceptibility profiles on 43 clinical isolates of A. baumannii.
AK, amikacin; GN, gentamicin; IMP, imipenem; MEM, meropenem; CAZ, ceftazidime; CXM, cefuroxime; CRO, ceftriaxone; AM, ampicilin; AmC, amoxicllin/ clavulanic acid (Augmentin); CIP, ciprofloxacin; NOR, norfloxacin; SXT, trimethoprim/Sulfamethoxazole; TE, tetracycline; C, chloramphenicol; and TGC, tigecycline
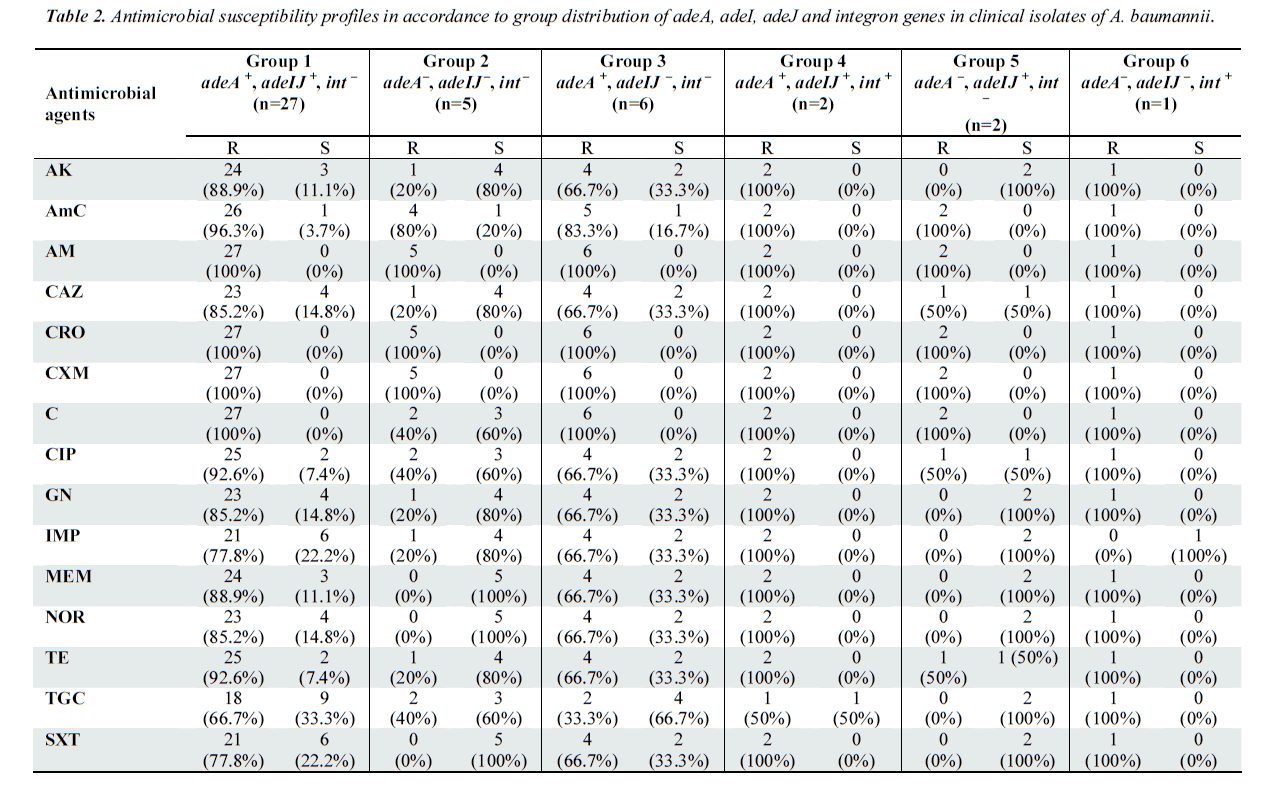
In this study, four RND pump system genes (adeA, adeI, adeJ, adeY) and integrons were chosen as the target gene. The specificity of all primers used was confirmed with nucleotide sequencing of generated PCR product (data not shown). From the results obtained, the distribution of RND pump genes and integrons were classified into six groups (Table 2). All of the clinical isolates were screened negative for adeY, indicating its non-existence in A. baumannii samples. The PCR results also revealed that 29 samples (67.4%) carried adeA, adeI as well carried adeJ genes, indicating that AdeABC and AdeIJK could coexist in A. baumanii [7,21].
Out of these 29 samples, a total of 27 (93.1%) samples did not harbor any integrons (Group 1), but this group displayed high resistance towards the antibiotics tested especially ampicillin, ceftriaxone, cefuroxime, chloramphenicol. The isolates classified under Group 3 and 5, which carried only one type of RND efflux genes showed reduced resistance to most of the antibiotics as compared to isolates from Group 1. This proved that isolates with the co-existence of both RND efflux pumps were significantly more resistant to all of the tested antibiotics.
Group 4 consisted of the two samples, which were positive for all adeA, adeI, adeJ genes and integrons. As observed in Table 2, the presence of the genes in these sam ples significantly increased the resistance towards all antibiotics tested significantly. Both the isolates in this group demonstrated resistance to all antibiotics, except for tigecycline. Our study revealed that adeA was the most prevalent RND pump, constituting to 81.4% (35/43) of the clinical isolates and this was followed closely by adeIJ which constituted 72.1% (31/43).
Only three out of 43 isolates (9.38%) was detected as class 1 integron positive: two isolates were placed in Group 4, while another was placed in Group 6. Interestingly, these samples exhibited much higher resistance to almost all of the antibiotics tested. PCR amplification of gene cassettes of class 1 integrons successfully generated amplicons with sizes of 2.3 kb. Nucleotide sequencing confirmed the presence of gene cassette aacA4-catB8- aadA1 in all three integron-positive isolates of A. baumannii.
Group 2 comprises of 11.6% of the clinical isolates, which did not carry any of the adeA, adeIJ or integron genes. Other than the penicillins (i.e. augmentin and ampicilin) and the cephalosporins (i.e. ceftriaxone and cefuroxime), it was observed that more than 60% of the isolates in this group were sensitive to the antibiotics, which harbored at least one of the genes tested. The results here confirm the role played by RND genes and integrons in bacterial multidrug resistance.
Discussion
In this study, adeA gene is shown to be the most prevalent in our clinical isolates. adeA gene encodes for one of the proteins that make up the tripartite system of efflux pump AdeABC. The finding is consistent with several studies, which showed the high prevalence of AdeABC amongst A. baumannii strains [22,23]. Both adeI and adeJ genes also encode for component proteins of efflux pump AdeIJK, and they are co-transcribed simultaneously [11]. Here, adeI and adeJ were proven to be highly prevalent in A. baumannii isolated from Malaysian hospitals, and they were always detected together in the isolates. The organization of three adjacent adeI, adeJ and adeK genes suggests that AdeIJK formed an operon that is similar to AdeABC efflux system [7]. Therefore, the results conformed to previous studies [7,11].
A high percentage of the isolates also harbored both adeA and adeIJ genes, which indicates the co-existence of AdeABC and AdeIJK efflux pumps system in A. baumannii. This is in line with other studies whereby majority of the isolates were positive for the presence of both groups of RND efflux pump and their roles in resistance development [16]. As expected, the co-existence of adeA and adeIJ genes greatly increased the resistance profile of the isolates, and thus, signifying their role in antibiotic resistance development [11]. This study also showed the increasing resistance toward tigecycline in this region, and thus conforming to recent studies [24,25].
Although not extensively distributed, the discovery of class 1 integrons in three of our clinical isolates is still an interesting find. The existence of the integron alone in the isolate (Group 6) rendered the isolate resistance to 14 out of 15 of the antibiotics tested. Similar to most studies, only class 1 integrons were detected here. Class 1 integrons are by far the most common class of integrons in clinical isolates of Gram-negative bacteria [15,19], including A. baumanii [26-29]. Gene cassette array showed the presence of a 2.3 kb cassette array, aacA4-catB8- aadA1, in all the integron-positive A. baumannii isolates. This array is particularly common in A. baumannii worldwide [29-31]. The high percentage of aacA4-catB8- aadA1 existence in class 1 integrons implicates that this cassette is the prevalent one among the A. baumannii isolates and the epidemiological relationship to other A. baumannii strains worldwide. Further studies are required to determine the clonal relationship with the A. baumannii strains reported in other countries, and also to determine if there are mutations occurring inside the gene cassettes that affect cassette gene expression.
In conclusion, the present work gives a brief description on the distribution of RND efflux pumps, integrons and their associated cassette arrays in A. baumannii isolates in Malaysia. A. baumannii isolates in this study demonstrate the frequent use of RND efflux pump genes rather than the acquisition of mobile gene cassette using site-specific recombination integrons.
Acknowledgement
We are grateful to Hospital Raja Permaisuri Bainun Ipoh, Gribbles® Pathology Sdn. Bhd, KPJ Ipoh Specialist Hospital and Hospital Pantai Putri Ipoh for their support of this study. This work was supported by Malaysia Toray Science Fund (MTSF) (4392/K01) and the Department of Biomedical Science, UTAR.
References
- Villa J, Marti S, Sanchez-Cespedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother 2007; 59: 1210-1215.
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global Challenge of Multidrug-Resistant Acinetobacter baumannii. Antimicrob Agents and Chemother 2007; 52(10): 3471-3484.
- Pittet D, Harbarth S, Ruef C, Francioli P, Sudre P, Pétignat C, Trampuz A, Widmer A. Prevalence and risk factors for nosocomial infections in four university hospitals in Switzerland. Infect Control Hosp Epidemiol 1999; 20(1): 37-42.
- Peleg AY, Adams J, Paterson DL. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother 2007; 51(6): 2065-2069.
- Wong EW, Yusof MY, Mansor MB, Anbazhagan D, Ong SY, Sekaran SD. Disruption of adeB gene has a greater effect on resistance to meropenems than adeA gene in Acinetobacter spp. isolated from University Malaya Medical Centre. Singapore Med J 2009; 50(8): 822-826.
- Magnet S, Courvalin P, Lambert T. Resistancenodulation-division type efflux pump in aminoglycoside resistance in Acinetobacter baumannii Strain BM4454. Antimicrob Agents Chemother 2001; 45(12): 3375-3380.
- Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two component system. Antimicrob Agents and Chemother 2004; 48(9): 3298-3304.
- Coyne S, Guigon G, Courvalin P, Perichon B. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents and Chemother 2010; 54(1), 333-340.
- Chau SL, Chu YW, Houang ETS. Presence of active efflux systems AdeABC, AdeDE and AdeXYZ in different Acinetobacter genomic DNA groups. J. Med Microbiol 2006; 55(4): 477-478.
- Wieczorek P, Sacha P, Hauschild T, Zorawski M, Krawczyk M, Tryniszewska E. Multidrug resistant Acinetobacter baumannii- the role of adeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cyto 2008; 46(3): 257-267.
- Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents and Chemother 2008; 52(2): 557-562.
- Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific geneintegration functions: integrons. Molecular Microbiology 1989; 3: 1669-1683.
- Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs 2004; 64: 159-204.
- Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol 2006; 4: 608–620.
- Turton JF, Kaufmann ME, Glover J, Coelho JM, Warner M, Pike R, Pitt TL. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J Clin Microbiol 2005; 43: 3074–3082.
- Lin L, Ling BD, Li XZ. Distribution of the multidrug efflux pump genes, adeABC, adeDE, and adeIJK, and class 1 integron genes in multiple-antimicrobialresistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int. J. Antimicrob Agents 2009; 33(1): 17-32.
- Cheong HT, Ho WY, Choo QC, Chew CH. β-lactamase gene blaSHV detected in bacteria isolated from retail sushi in Kampar, Malaysia. Biomedical Research-India 2014; 25(1): 25-31.
- White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother 2001; 45: 2658-2661.
- Kor SB, Choo QC, Chew CH. New integron gene arrays from multiresistant clinical isolates of members of the Enterobacteriaceae and Pseudomonas aeruginosa from hospitals in Malaysia. J Med Microbiol 2013; 62: 412–420.
- Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 1995; 39: 185-191.
- Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. New insights into Acinetobacter baumannii pathogenesis revealed by high density pyrosequencing and transposon mutagenesis. Genes Dev 2007; 21: 601-614.
- Chu YW, Chau SL, Houang ETS. Presence of active efflux systems AdeABC, AdeDe and AdeXYZ in different Acinetobacter genomic DNA groups. J. Med Microbiol 2006; 55: 477-478.
- Yoon EJ, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother 2013; 57(7): 2989-2995.
- Rumbo C, Gato E, López M, Ruiz de Alegría C, Fernández-Cuenca F, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual A, Bou G, Tomás M. Contribution of Efflux Pumps, Porins, and β-Lactamases to Multidrug Resistance in Clinical Isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 2013; 57(11): 5247- 5257
- Horn.s e y M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother 2010; 65(8): 1589-1593.
- Gombac F, Riccio ML, Rossolini GM, Lagatolla C, Tonin E, Monti-Bragadin C, Lavenia A, Dolzani L. Molecular characterization of integrons in epidemiologically unrelated isolates of Acinetobacter baumannii from Italian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob Agents Chemother 2002; 46(11): 3665-3668.
- Koeleman JG, Stoof J, Van Der Bijl MW, Vandenbroucke-Grauls CM, Savelkoul PH. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J Clin Microbiol 2001; 39(1): 8-13.
- Çıçek AÇ, Düzgün AÖ, Saral A, Kayman T, Çızmecı Z, Balcı PÖ, Dal T, Fırat M, Tosun İ, Alıtntop YA, Çalışkan A, Yazıcı Y, Sandallı C. Detection of class 1 integron in Acinetobacter baumannii isolates collected from nine hospitals in Turkey. Asian Pac J Trop Biomed 2013; 3(9): 743-747.
- Lin MF, Liou ML, Tu CC, Yeh HW, Lan CY. Molecular epidemiology of integron-associated antimicrobial gene cassettes in the clinical isolates of Acinetobacter baumannii from northern Taiwan. Ann Lab Med 2013; 33(4): 242-247.
- Gu B, Tong M, Zhao W, Liu G, Ning M, Pan S, Zhao W. Prevalence and characterization of class 1 integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol 2007; 45(1): 241-243.
- Gallego L, Towner KJ. Carriage of class 1 integrons and antibiotic resistance in clinical isolates of Acinetobacter baumannii from northern Spain. J Med Microbiol 2001; 50: 71-77.