- Biomedical Research (2015) Volume 26, Issue 1
Detection of ?-lactamase genes, ERIC-PCR typing and phylogenetic groups of ESBL producing quinolone resistant clinical Escherichia coli isolates.
Suleyman Durmaz1, E. Banu Buyukunal Bal2*, Munevver Gunaydin2, Erkan Yula3, Duygu Percin41Department of Medical Microbiology, School of Medicine, Mevlana University, Konya, Turkey
2Department of Biology, Faculty of Science and Letters, Kahramanmaras Sutcu Imam University, Kahramanmaras, Turkey
3Department of Medical Microbiology, School of Medicine, Katip Celebi University, Izmir, Turkey
4Department of Medical Microbiology, School of Medicine, Erciyes University, Kayseri, Turkey
- *Corresponding Author:
- E. Banu Buyukunal Bal
Kahramanmaras Sutcu Imam University
Faculty of Science and Letters
Department of Biology
Avsar Campus, 46050, Turkey
Accepted date: September 12, 2014
Abstract
Escherichia coli are one of the most important agents in clinical settings. Treatment of E. coli infections are being complicated due to emergence of resistant bacteria which limits the choices of treatment. The extended spectrum of β-lactamase (ESBL) producing and quinolone resistant E. coli isolates (n= 109) have been isolated from various clinical specimens belonging to both out-coming and in-coming patients of the Erciyes University Medical School Hospital of Turkey. Occurrence of genes encoding major type of β-lactamases (blaCTX-M, blaSHV and blaTEM) was investigated by using PCR among the isolates. ERIC-PCR typing was performed on the isolates. Moreover, phylogenetic groups were determined. The blaCTX-M was detected at the highest frequency (93%; 101/109), while the blaSHV and blaTEM were present in 65% (71/109) and 49% (56/109), respectively. Sequencing of β-lactamase genes from several randomly selected isolates revealed that CTX-M-15 is the only CTX-M type enzyme, while there is variation in possessing SHV and TEM types of enzymes. Isolates harboring three bla genes were 32%. ERIC-PCR analyses, based on two different primers, demonstrated 5 to 6 clonal groups for 42 isolates. Phylo-grouping of all isolates was shown that majority of isolates was belonging to B2 (46%) and D (31%) major groups. The detection of genes encoding major β-lactamases (blaCTX-M, blaSHV and blaTEM) was very high in ESBL-producing quinolone resistant E. coli phenotypic isolates. It indicates that the molecular data was congruent with the phenotypic selection results.
Keywords
E. coli, ERIC-PCR, ESBL, Quinolone.
Introduction
Members of Enterobacteriaceae are important causative agents for both hospital and community acquired infections. Among those, Escherichia coli and Klebsiella pneumoniae are the most frequently encountered. Treatment of infections resulting from those agents has been hampered due to emergence of antibiotic resistant bacteria [1]. Although there are variety of ways exist for emerging antibiotic resistant bacteria, persistent and longterm use of antibiotics can be noticeable for bacterial resistance mechanisms for horizontal transfer of resistance determinants and their associated genetic elements [1,2]. In a similar way, a group of antibiotics, called β-lactams, have been used to a great extent in clinical and veterinary practices.
β-lactamases produced by most of the gram negative bacteria can hydrolyze β-lactams, therefore they are responsible from resistance [1]. Various β-lactamases with different substrate specificities and molecular characteristics have been characterized from different members of Enterobacteriaceae and from non-enteric organisms, such as Acinetobacter spp. up to date [3-5]. Among those enzymes, some are termed as an extended spectrum of β-lactamases (ESBLs) and derived from parent types of β-lactamases, such as SHV-1 and TEM-1. Those can differ from their originated enzymes with their ability for hydrolyzing extended-spectrum cephalosporins and are generally inhibited by lactamase inhibitors, such as clavulanic acid. Moreover, recent reports indicated that ESBL producing isolates have been occurred at increasing rate in worldwide, since ESBL’s discovery [6]. This increase is most dramatic in countries where antibiotics are used irregularly. As a consequence, ESBL producing clinical isolates either Enterobacteriaceae members or other gram negative bacteria (Pseudomonas aeruginosa and Acinetobacter baumanii) have been previously identified from Turkey [7-9]. Most of the time, quinolones are the first choice against infections caused by ESBL producing organisms; however, genes conferring resistance to both β-lactams and quinolones are frequently transferred on the same plasmids. In the case of co-existence of resistance, efficiency of quinolones is also limited for the successful treatment. Therefore, resistance against both types of antibiotics should be investigated together in clinical isolates. The aims of this study were to determine the occurrence of major β- lactamase producing genes in ESBL producing quinolone resistant clinical E. coli isolates as an indication of recent risk status of ESBL producing isolates, characterize β- lactamase enzymes by sequencing, detect genetic diversity on a collection of isolates, and determine the phylogenetic groups of isolates.
Materials and Methods
Sample collection and isolation of E. coli
The used E. coli isolates (n= 109) were phenotypically characterized as ESBL producing and quinolone resistant. The study protocol was approved by the Ethical Committee of Erciyes University Medical School. All clinical samples (76 urine, 12 blood, 9 wound, 4 drainage, 3 endo-tracheal aspirate (ETA), 2 naso-tracheal aspirate (NTA), 1 abscess, 1 peritoneum, 1 intra-abdominal secret) were collected from patients with community or hospital acquired E. coli infections from the Erciyes University Medical School, Gevher Nesibe Research Hospital in Kayseri, Turkey. Classification of the infections was performed based on the hospital records. If an infection has been seen after 72 hours of hospitalization or 10 d after leaving from the hospital, it was classified as hospital acquired [10]. K. pneumoniae (ATCC 70603) and E. coli (ATCC 25922) strains were used as positive and negative controls for ESBL detection. Clinical samples which are suspected to have infection were cultured on blood and Eozin Metilen Blue (EMB) (Oxoid, England) agar. After incubation, gram negative bacteria were identified at species level based on standard biochemical tests.
Antibiotic susceptibility testing
Detection of ESBL producing isolates was performed with double disk synergy test using following antibiotic disks: amoxycillin/clavulanic acid (30 μg), aztreonam (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg) and cefpodoxime (10 μg) (Oxoid, UK) (CLSI, 2011). Briefly, inoculums were prepared in sterile saline solutions from grown cultures. When 0.5 McFarland turbidity value was obtained for each bacterial inoculums, they were inoculated onto Mueller Hinton Agar (Oxoid, UK) by sterile cotton swab. Then, antibiotic disks were placed in a way that amoxycillin/clavulanic acid was at the center and the others around it that was far from each other (24 mm). Subsequently, they were incubated at 37ºC for 18-20 hours. Enlargement of zones at the sites facing to amoxycillin/clavulanic acid disk was considered as ESBL positive. Resistance to quinolones and other antibiotics was determined with Kirby-Bauer disk diffusion test using ciprofloxacin disk (5 μg) as described by CLSI11. E. coli ATCC 25922 was used as quality control strain. Results were interpreted according to CLSI criteria [11].
Detection of β -lactamase genes
ESBL producing E. coli isolates were grown on EMB agar. Colonies were then inoculated to TSB and grown at 37oC for 18-20 hours. Grown cultures (1 ml) were used to obtain cellular DNA samples. Following to centrifugation for 10 min at 12000 g, pellets were suspended in 200 μl of TE buffer. Samples were then incubated at 95oC for 10 min and were re-centrifuged. Supernatants (5 μl) were used as template in PCR amplifications. All E. coli isolates were screened for the presence of CTX-M, TEM, and SHV encoding genes using previously described primer sets [12,13]. Sequence analysis was performed on PCR products from randomly selected few isolates to identify specific β- lactamase genes (blaTEM, blaSHV, blaCTX-M) (Refgen Biotechnology Co., Turkey). Nucleotide sequences were compared using the BLAST search tool (http://www.ncbi.nlm.nih.gov/Blast.cgi).
ERIC-PCR typing
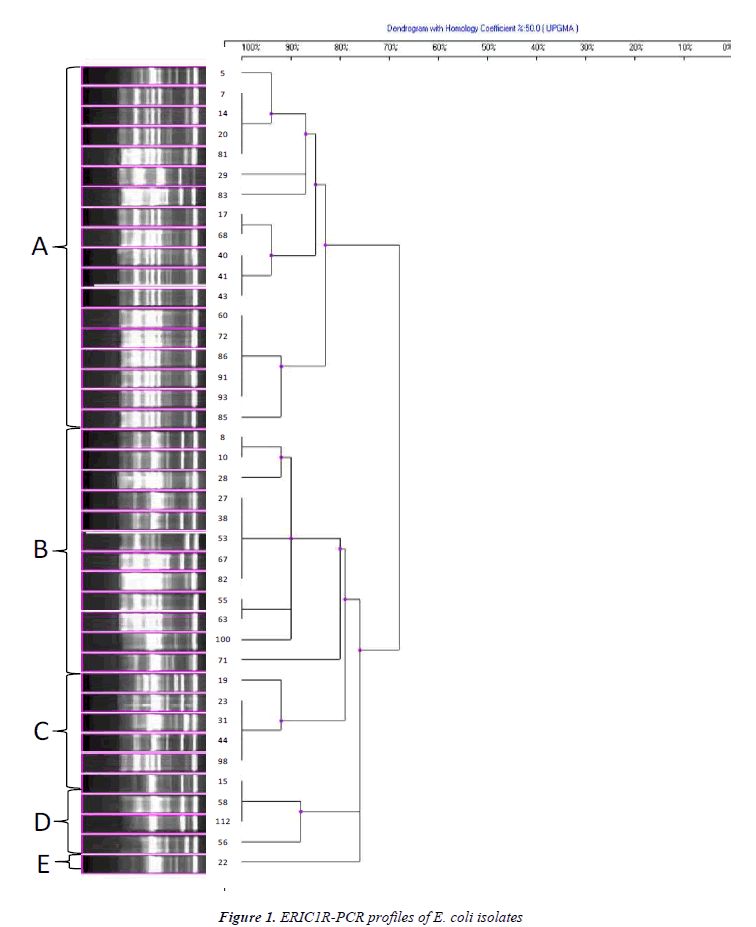
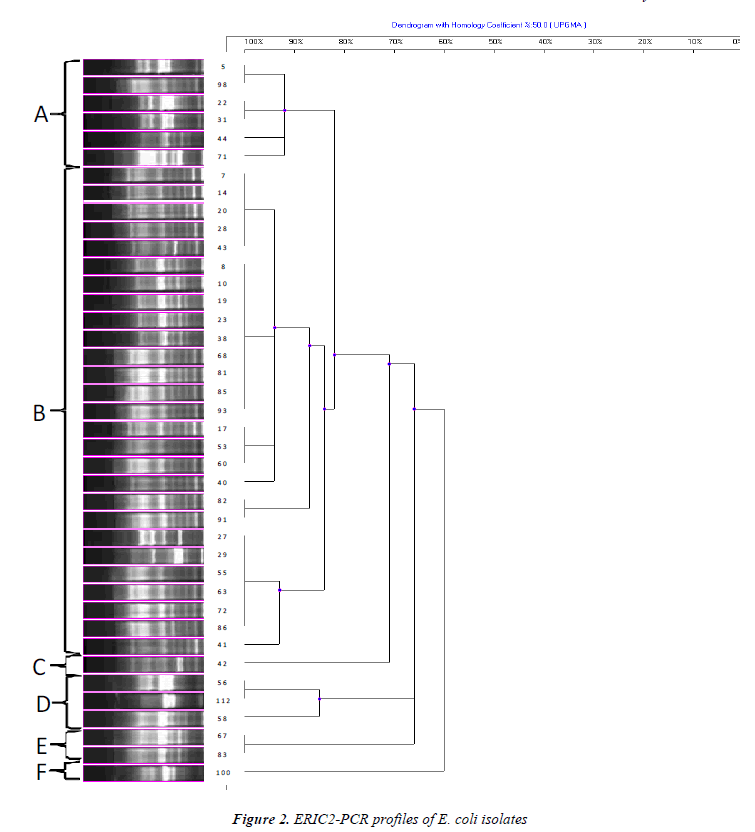
Genomic DNAs belonging to 42 isolates, used for ERICPCR typing, were extracted by commercial kit (Favorgen, Taiwan). The primers used for ERIC-PCR typing were as follows: ERIC-1R (5'-AAGCTCCTGGGGATTCA-3') and ERIC-2 (5'-AAGTAAGTGACTGGGGTGAGCG-3') [14,15]. Each PCR amplification was performed on 50 μl of total volume containing 5 μl of target DNA, 0.2 mM dNTP, 50 pmol of each primer, 5 μl of 10X PCR buffer solution (500 mM KCl, 100 mM Tris-HCl, pH 9.0), 2.5 mM MgCl2 and 2 units of Taq DNA Polymerase (Fermentas).
PCR conditions were modified as following: denaturation at 95oC for 2 min, 40 cycles with denaturation at 92oC for 30 sec, annealing at 50oC for 1 min and polymerization at 72oC’ for 8 min. After reactions were completed, final extension was performed at 72oC for 16 min [16]. PCR products were separated by electrophoresis on 1% agarose gels and visualized using a gel documentation system (Vilber Lourmat, France) after staining with ethidium bromide. PCR profiles obtained with ERIC primers were used for computation of Dice similarity coefficient values [17]. Subsequently, those values were used to construct dendograms using Unweighted Pair Group Method with Arithmetic Mean (UPGMA). Isolates with similarity >80% were considered to be within a group.
Phylogenetic grouping
Three sets of primers (ChuA.1 (5'-GACGAACC AACGGTCAGGAT-3'), ChuA.2 (5'-TGCCGCCAGTA CCAAAGACA-3'), YjaA.1 (5'-TGAAGTGTCAGGAG ACGCTG-3'), YjaA.2 (5'-ATGGAGAATGCGTTCCTC AAC-3'), TspE4C2.1 (5'-GAGTAATGTCGGGGCAT TCA-3'), TspE4C2.2 (5'-CGCGCCAACAAAGTATT AC G-3') were used in multiplex PCR method as described previously [18]. E. coli ATCC 25922 belonging to B2 phylo-group was used as a control strain during PCR amplification. PCR products were separated on 1% agarose gels and visualized on gel documentation system (Vilber Lourmat, France) to detect three bands with 279, 211 and 152 bp sizes, respectively.
Results and Discussion
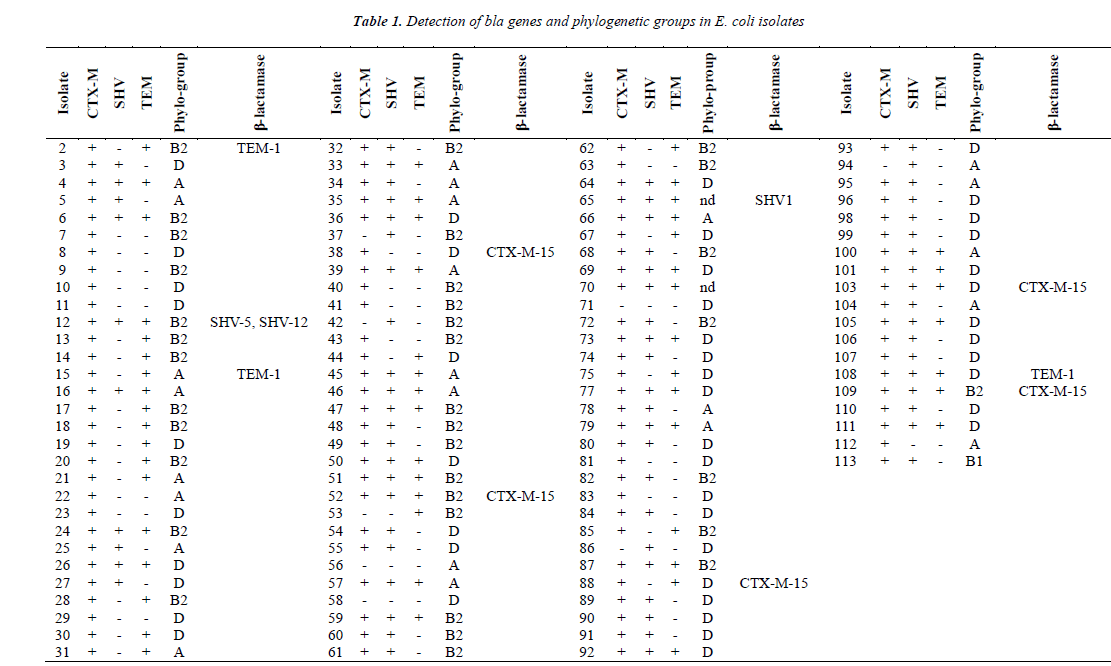
PCR detection results gave rise to different bla genes encoding major ESBLs such as TEM, SHV, CTX-M (Table 1). Although prevalence of CTX-M encoding gene in ESBL producing E. coli from Turkey varies between 54.5-100%, its prevalence was found to be the highest amongst bla genes. The most drastic change in this study was observed for the prevalence of the blaSHV gene as compared to the results from other studies [8,13]. However, a quite similar rate (74.3%) for the presence of blaSHV was reported previously. All ESBL producing and quinolone resistant E. coli isolates examined in this study have been shown to contain at least a single bla gene (CTX-M, SHV, TEM) by PCR. Moreover, 32% (35/109) of isolates were shown to contain all bla genes for CTX-M, TEM and SHV production. This result might be expected considering the carriage of CTX-M and TEM producing genes on the same plasmid.
CTX-M encoding gene has been most frequently detected from community acquired E. coli infections, while TEM and SHV encoding genes have been detected mostly in hospital acquired infections [19,20]. However, the blaCTX-M gene in the present study was detected and distributed in almost all isolates acquired from the community and hospital. This result indicates both community- and hospital-acquired isolates might be equally important for the transmission of this gene. CTX-M producing isolates were commonly found to be multiple antibiotic resistant. For instance, CTX-M producing isolates were resistant to fluoroquinolones as genes conferring resistances against other type of antibiotics are located on the same plasmid [21].
Similar to CTX-M producing isolates, TEM and SHV types of ESBLs producing isolates are often resistant to aminoglycosides, tetracycline and sulphonamides. Consistently, all E. coli isolates used in this study were phenotypically resistant to ciprofloxacin, majority of which carrying the blaCTX-M gene also possesses the blaSHV and blaTEM genes. A recent study conducted at the same hospital indicated that quinolone resistance among ESBL-producing E. coli isolates was higher than non-ESBL producers [22]. This result was consistent with the results obtained from Iran, which is geographically close to Turkey [23]. Similarly, in most European countries the prevalence of E. coli resistance to fluoroquinolones, the third-generation cephalosporins, and aminoglycosides increase every year [24].
Trimethoprim/sulfamethoxazole, aminoglycosides, fluoroquinolones, β-lactam/β-lactamase inhibitor combinations, and carbapenems represent valuable therapeutic options against ESBL producing organisms. However, there have been increasing reports of a decrease in susceptibility to trimethoprim/ sulfamethoxazole, aminoglycosides and fluoroqui-nolones [25,26].
Therefore, antibiotics resistance incidences were investigated here for several antibiotics belonging to each group above. As a result, 23.1% (25/108) resistance to amoxicillin-clavulanic acid, 7% (7/100) resistance to piperacillin-tazobactam, 10.6% (11/104) resistance to amikacin, 0.9% (0/107) resistance to imipenem and 14% (12/86) to netilmicin were found. This result shows that imipenem and piperacillin-tazobactam could be still valuable for treatment of infections caused by both quinolone resistant and ESBL-producing organisms. Moreover, amoxicillin-clavulanic acid resistance was relatively high and further analysis is required to determine the exact resistance mechanisms in resistant isolates.
Although variants of CTX-M gene have been detected at increasing rate from community acquired infections from different geographical sites belonging to different species, CTX-M-15 is the most prevalent [27]. This type is related to ST131 pandemic clone which contribute the rapid dissemination of this ESBL. In this study, randomly selected isolates having blaCTX-M gene have been sequenced and CTX-M-15 has been the only type among detected confirming the results of the previous study [28]. In another study, CTX-M-15 was detected in 53% of ESBL positive E .coli isolates from communityacquired urinary tract infections (UTIs) infections in Western Turkey and all CTX-M-15 isolates shared the same PFGE profiles with ST131 clone indicating that they belong to this clone [29]. Another recent study reported that other types of CTX-M, such as CTX-M-1 and CTX-M-3 type have been detected in low frequencies among ESBL-producing E. coli UTI related isolates along with predominant type of CTX-M-15 [13].
Molecular studies towards characterization of TEM and SHV types of β-lactamases in clinical isolates belonging to Enterobacteriaceae from Turkey were few. The results of those studies indicated that SHV-1, SHV-2, SHV-5 and SHV-12 are the detected types in Turkey and SHV-2 has been found widely. A study indicated that SHV-12 was the one reported for first time from clinical E. coli isolates from Turkey [8]. In the present study, SHV-1, SHV-5 and SHV-12 types were detected in two isolates based on partial sequencing results of randomly selected isolates.
However, two blaSHV genotypes (SHV-5 and SHV-12) have been detected for isolate 12 based on the partial sequencing result of blaSHV gene. Therefore, addressing the presence of different SHV enzymes may need further research.
The genetic diversity among ESBL producing quinolone resistant 42 clinical E. coli isolates was successfully recovered by ERIC-PCR (Figures 1 and 2). Clustering of isolates based on ERIC-PCR profiles revealed that presence of diverse clonal groups indicated transmission among members of separate groups. Although grouping of isolates were slightly changed in the two dendograms based on different ERIC primers (ERIC-1R and ERIC-2), positioning of most isolates were in agreement with two dendograms. For instance, isolate 8 and 10 grouped together in both dendograms, similarly isolates 55 and 63 were located on the same sub cluster. ERIC-PCR analysis based on different primers gave rise to similar number of major groups, however distinction of certain isolates have been achieved more efficiently when different primers were used. For instance, isolate 5 has been differentiated with ERIC-1R primer, however ERIC-2 primer was better for differentiation of isolate 41. Some isolates used in both analyses had distinct clonal profile, such as isolates 71 and 100.
Detection of more isolates belonging to D and B2 phylogroup agrees with the results of previous studies, indicating that the isolates of B2 and D phylo-groups contain more virulence factors than the isolates of A and B1 phylo-groups [30,31]. Recent studies also highlighted that these phylo-groups differ with ecological niches, lifehistory and some characteristics, antibiotic-resistance profiles and growth rate of bacteria [32]. In our study, CTX-M-15 enzyme was more common in isolates belonging to B2 and D phylo-groups. This was similar to the results of another study which indicated CTX-M producers, especially CTX-M-15 ones, and was significantly associated with phylogenetic group B2 [33].
In conclusion, the occurrence of genes encoding major β- lactamases (blaCTX-M, blaSHV and blaTEM) was very high in ESBL-producing quinolone resistant E. coli phenotypic isolates. Moreover, results of ERIC-PCR analysis demonstrated the presence of 5 to 6 clonal groups. Few isolates responsible for both community and hospital acquired infections have conserved banding patterns in ERIC analyses, indicating rapid dissemination among clonal groups. In addition, there need to be precise epidemiological investigations for monitoring isolates more frequently. These analyses will also aid in differentiating strains causing sporadic and (or) outbreak infections along with selection of efficient treatment options for preventing transmission.
References
- Livermore DM. Beta-lactamases in laboratory and clinical resistance. Clin Microbiol Rev., 1995; 8: 557-584.
- Bean DC, Livermore DM, Hall LM. Plasmids imparting sulfonamide resistance in Escherichia coli: implications for persistence. Antimicrob Agents Chemother., 2009; 53: 1088-1093.
- Bush K., Jacoby GA, Medeiros AA. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother., 1995; 39: 1211-1233.
- Naas T, Philippon L, Poirel L, Ronco E, Nordmann P. A SHV-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother., 1999; 43: 1281-1284.
- Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev., 2001; 48: 933-951.
- Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill., 2008; 13: 19-29.
- Hall LMC, Livermore DM, Gur D, Akova M, Akalin HE. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother., 1993; 37:1637-1644.
- Tasli H, Bahar IH. Molecular characterization of TEM- and SHV-derived extended-spectrum beta-lactamases in hospital based Enterobacteriaceae in Turkey. Jpn J Infect Dis., 2005; 58: 162-165.
- Vahaboglu H, Budak F, Kasap M, Gacar G, Torol S., Karadenizli A, Kolayli F, Eroglu C. High prevalence of OXA-51-type class D β-lactamases among ceftazidime resistant clinical isolates of Acinetobacter spp.: co- existence with OXA-58 in multiple centers. J Antimicrob Chemother., 2006; 258: 537-542.
- Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections. Am J Infect Control., 1988; 16: 128-140.
- Clinical and Laboratory Standards Institute. Performa- nce Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. CLSI docu- ment (M100-S21), CLSI, 2011; Wayne PA.
- Naiemi N, Duim B, Savelkoul PHM, Spanjaard L, de Jonge E, Bart A, Vandenbroucke-Grauls CM., de Jong MD. Widespread transfer of resistance genes between bacterial species in an intensive care unit: implications for hospital epidemiology. J Clin Microbiol., 2005; 43: 4862-4864.
- Celik AD, Yulugkural Z, Kuloglu F, Eroglu C, Torol S, Vahaboglu H, Akata F. CTX-M Type extended spectrum β-lactamases in Escherichia coli isolates from community acquired upper urinary tract infections at a university in the European part of Turkey. J Microbiol Immunol Infect., 2010; 43: 163-167.
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA Sequences in Eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res., 1991; 19: 6823-6831.
- Savli H, Karadenizli A, Kolayli F, Gundes S, Ozbek U, Vahaboglu H. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microbiol., 2003; 52: 403-408.
- Decre D, Burghoffer B, Gautier V, Petit JC, Arlet G. Outbreak of multi-resistant Klebsiella oxytoca involving strains with extended spectrum β-lactamases and strains with extended-spectrum activity of the chromosomal β-lactamase. J Antimicrob Chemother., 2004; 54: 881-888.
- Lee DR. Measures of the amount of ecologic association between species. Ecology, 1945; 26: 297-302.
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol., 2000; 66: 4555-4558.
- Paterson DL, Bonomo RA. Extended-spectrum beta- lactamases: a clinical update. Clin Microbiol Rev., 2005; 18: 657-686.
- Pitout JD, Laupland KB. Extended-spectrum beta- lactamase-producing Enterobacteriaceae: an emerging public health concern. Lancet Infect Dis., 2008; 8: 159-166.
- Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, Pearson A, Harry S, Leach JB, Loughrey A, Lowes JA, Warren RE, Livermore DM. Community and hospital spread of Escherichia coli producing CTX-M extended spectrum beta-lactamases in the UK. J Antimicrob Chemother., 2004; 54: 735-743.
- Saglam D, Durmaz S, Kilic H, Atalay MA, Ercal BD, Sarli S, Percin D. The frequency of extended-spectrum beta-lactamases and antibiotic resistance patterns in Escherichia coli strains isolated from blood cultures. Ankem Derg., 2011; 25: 250-255.
- Pakzad I, Ghafourian S, Taherikalani M, Sadeghifard N, Abtahi H, Rahbar M., Mansory Jamshidi N. qnr prevalence in extended spectrum Beta-lactamases (ESBLs) and none-ESBLs producing Escherichia coli isolated from urinary tract Infections in central of Iran. Iran J Basic Med Sci., 2011; 14: 458-464.
- Colodner R, Kometiani I, Chazan B, Raz R. Risk factors for community-acquired urinary tract infection due to quinolone-resistant E. coli. Infection 2008; 36: 41-45.
- Lautenbach E, Strom BL, Bilker WB, Patel JB, Edelstein PH, Fishman NO. Epidemiological investigation of fluoroquinolones resistance in infections due to extended-spectrum beta lactamase- producing Escherichia coli and Klebsiella pneumoniae. Clin Infect Dis., 2001; 33: 1288-1294.
- Koksal F, Ak K, Kucukbasmaci O, Samasti M. Prevalence and antimicrobial resistance patterns of extended-spectrum beta lactamase- producing Escherichia coli and Klebsiella pneumoniae isolated from blood cultures in an Istanbul University Hospital. Chemotherapy 2009; 55: 293-297.
- Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol., 2006; 9: 466-475.
- Gonullu N, Aktas Z, Kayacan CB, Salcioglu M, Carattoli A, Yong DE, Walsh TR. Dissemination of CTX-M-15 beta-lactamase genes carried on Inc FI and FII plasmids among clinical isolates of Escherichia coli in a university hospital in Istanbul, Turkey. J Clin Microbiol., 2008; 46: 1110-1112.
- Yumuk Z, Afacan G, Nicolas-Chanoine M, Sotto A, Lavigne J. Turkey: A further country concerned by community-acquired Escherichia coli clone O25 ST131 producing CTX-M-15. J Antimicrob Chemo- ther., 2008; 62: 284-288.
- Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis., 1998; 177: 642-650.
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis., 2000; 181: 261-272.
- Gordon DM. The influence of ecological factors on the distribution and genetic structure of Escherichia coli. In: Neidhardt FC et al., editors, Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, DC, 2004.
- Mnif B, Harhour H, Jdidi J, Mahjoubi F, Genel N, Arlet G, Hammami A. Molecular epidemiology of extended-spectrum beta-lactamase-producing Escheric- hia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiology 2013; 13: 147-155.