Research Article - Biomedical Research (2017) Volume 28, Issue 7
Decreased expression of BECN1 mRNA is associated with lupus nephritis
Xiong-Yan Luo1#, Ming-Hui Yang2#, Qin Wei3#, Kuen Li3, Yi Liu1 and Guo-Hua Yuan2*1Department of Rheumatology, West China Hospital, Sichuan University, Chengdu, Sichuan, China
2Department of Clinical Laboratory, Affiliated Hospital of North Sichuan Medical College, Nanchong, Sichuan, China
3Department of Rheumatology, the First People’s Hospital of Xinxiang, Xinxiang Medical University, Xinxiang, Henan, China
#These authors contributed equally to this work
- *Corresponding Author:
- Guo-Hua Yuan
Department of Clinical Laboratory
Institute of Rheumatology and Immunology
Affiliated Hospital of North Sichuan Medical College, China
Accepted on November 28, 2016
Abstract
This study aimed to investigate the mRNA expression of the autophagy-related gene BECN1 in Systemic Lupus Erythematosus (SLE) and to analyse the relationship of its expression with SLE clinical characteristics and disease activities. Quantitative polymerase chain reaction was performed to detect the mRNA expression levels of BECN1 in 79 SLE patients and 45 healthy controls. BECN1 was downregulated in the 79 patients with SLE (P<0.001), and those with downregulated BECN1 mRNA exhibited a significantly increased incidence of nephritis (85.3% vs. 64.4%, P=0.038). BECN1 mRNA downregulation, which may be related to deficiency in autophagosome formation, can promote the pathogenesis of SLE and nephritis.
Keywords
Autophagy-related gene, ATG6/BECN1, Lupus nephritis.
Introduction
Systemic Lupus Erythematosus (SLE) is an autoimmune disease characterized by the generation of nuclear-targeting autoantibodies, consequently resulting in autologous systemic damages [1]. Although inflammation and vasculitis induced by the deposition of immune complexes are considered the pathological basis of SLE, its precise mechanism is still unknown [2,3]. Autophagy is a lysosome-mediated metabolic process that degrades and reuses cytoplasmic components specifically in old or damaged organelles, thus playing an important role in maintaining intracellular homeostasis [4-6]. In the immune system, autophagy can eliminate intracellular nucleic acids and other wastes; thus, it is an important process in innate immune responses. In addition, autophagy participates in the intrathymic-positive and -negative selection of lymphocytes; it eliminates intracellular antigenic materials, processes them into antigenic polypeptides, and commits them to immune cells, thus playing vital roles in acquired immunity [7,8]. Current studies have indicated that the abnormal expression of autophagy genes may be related to the occurrence of autoimmune diseases such as Crohn's disease, multiple sclerosis, or rheumatoid arthritis [9-11].
Autophagy begins with the formation of a complex that initiates phagophore membrane formation, involving BECN1, class III phosphoinositide 3-kinases, Vacuolar Protein Sorting 34 (VPS34), and other autophagy proteins. BECN1 is a key protein in autophagy initiation and is considered the gatekeeper of autophagy. BECN1 mRNA has been confirmed to be upregulated in murine experimental autoimmune neuritis, indicating that changes in BECN1 expression are involved in the pathological process of autoimmune diseases [12,13].
It has been reported that the abnormal expressions of the autophagy genes ATG5, ATG7, LC3, HSPA, UVRAG, and IRGM are related to an imbalance of T-cell functions in patients with SLE [14], and this imbalance plays a key role in the pathogenesis of SLE. However, whether the expression of BECN1, the gatekeeper of autophagy, is altered in patients with SLE and related to the clinical manifestations of SLE remain unclear. Therefore, we determined BECN1 expression in patients with SLE to investigate its expression pattern and relevance to the clinical manifestations of SLE.
Materials and Methods
Subjects
A total of 79 inpatients and outpatients were enrolled from Xinxiang People's Hospital, the Affiliated Hospital of North Sichuan Medical College, and West China Hospital of Sichuan University from June 2011 to December 2012. The patients included 13 males and 66 females, ranging from 15 to 58 years with a mean age of 33.2 years. Disease duration ranged from 0.5 to 173 months with an average of 29.7 months. All patients met the classification criteria for SLE issued by the American Rheumatism Association (ARA) in 1997, and SLE Disease Activity Index (SLEDAI) were used to determine disease activity. In addition, 45 gender- and age-matched healthy volunteers were selected as the control group, including 5 males and 40 females, ranging from 16 to 60 years with a mean age of 30.4 years. This study followed the ethical standards, and was approved by the Ethics Committee of our department, and all study subjects signed the informed consent.
Reagents and equipment
The human lymphocyte separation medium Ficoll-Hypaque (Tianjin Haoyang Biological Manufacture Co., Ltd, Tianjinn China), the total RNA extraction reagent Trizol (Takara Co.), the ReverTra Ace qPCR RT Kit (BRK 100T), and the 7900 HT Fast Real-Time PCR System (ABI, USA) were used in this study.
Specimen collection and peripheral blood mononuclear cell (PBMC) separation
From each study subject, 5 ml of peripheral blood was collected early in the morning. PBMC was separated from the collected blood, using Ethylenediaminetetraacetic acid (EDTA)-K2 anticoagulant and Ficoll-Hypaque following the density gradient centrifugation method according to protocol. Monocytes were then removed according to the wall adherence method [14]. The cells obtained were then counted, and peripheral blood lymphocytes were collected for total RNA extraction.
Total RNA extraction
According to protocol, total RNA was extracted using Trizol reagent, dissolved in Diethyl Pyrocarbonate (DEPC)-treated deionized water, and stored at-80°C. UV spectrophotometry was performed to analyse its purity and concentration.
cDNA synthesis
cDNA was synthesized according to the instructions of the ReverTra Ace qPCR RT Kit. The oligo (dT) 18 Primer was selected, and 9 μL of RNA solution was reverse transcribed into cDNA and stored at-20°C.
Reverse transcription-polymerase chain reaction (RT-PCR)
The primers of the target gene (Gene ID: 8678) and the internal reference GAPDH (Gene ID: 2597) for RT-PCR were synthesized by BIONEER Co., (Pudong, Shanghai, China).
The total volume (20 μl) consisted of 8 μl of ddH2O, 9 μl of SYBR mixture, 0.5 μl of forward primer (10 μmol/L), 0.5 μl of reverse primer (10 μmol/L), and 2 μl of cDNA. PCR cycling conditions were 95°C for 30 s, 95°C for 5 s, 52°C for 5 s, and 72°C for 30 s, with a total of 40 cycles. The results of RT-PCR are expressed in cycle number (Ct), the point in the reaction at which the fluorescence signal in each tube reached the preset threshold; the greater the Ct value, the lower the amount of the starting template, and vice versa. The ΔCt value was obtained by subtracting the Ct value of the housekeeping gene GAPDH from the gene Ct value; the greater the ΔCt value, the lower the gene expression level. RQ=2-(ΔΔCt) was used to determine the mRNA expression level; the higher the RQ value, the higher the mRNA expression level [3]. The reaction was performed using the ABI 7900HT Fast Real-Time PCR System, and each run included a template-free control. All experiments with the specimens and controls were repeated to obtain an average value.
Statistical analysis
Measurement data are expressed as ͞x ± s. The comparison of measurement data between two groups was performed by t-test, comparison of count data was performed by Pearson’s Chi-square (χ2) test, and comparison of measurement data among multiple groups was performed by one-way ANOVA. All data were processed using the SPSS 11.5 software package with P<0.05 considered as statistically significant.
Results
RT-PCR
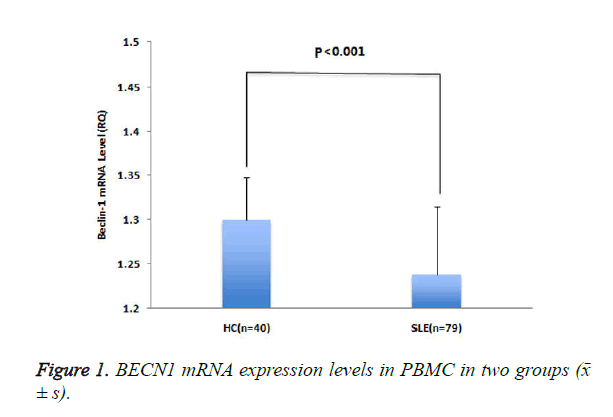
The expression level of BECN1 mRNA (RQ) in patients with SLE was lower than that in healthy controls (P <0.001) (Figure 1).
Correlations between BECN1 mRNA expression level and SLE disease activity
The results revealed a significant difference in the expression levels of BECN1 mRNA between patients with SLE and healthy controls. To determine whether this difference may affect SLE, we further analysed the correlations between BECN1 mRNA expression level and clinical manifestations of patients. The mean expression level of BECN1 mRNA in the control group (-2 SD, specifically RQ=1.207) was set as the normal lower limit, and RQ<1.27 was defined as BECN1 mRNA downregulation. The results showed that SLE patients with increased BECN1 mRNA downregulation had a higher disease activity score than healthy controls (8.06 ± 2.83 vs. 6.01 ± 2.56, P=0.004), and further analysis revealed that the expression level of BECN1 mRNA was negatively correlated with SLEDAI (r=-0.267, P=0.001).
Correlations between BECN1 mRNA expression level and clinical features of SLE
To investigate whether the expression level of BECN1 mRNA was related to certain clinical features of SLE, we further analysed and compared the clinical features and laboratory results of patients with SLE who had different BECN1 mRNA expression levels. Patients with SLE with downregulated BECN1 mRNA exhibited a significantly increased incidence of nephritis (85.3% vs. 64.4%, P=0.038) (Table 1).
| ATG6/Beclin-1 | RQ<1.207 (n=34) | RQ ≥ 1.207 (n=45) | P-value |
|---|---|---|---|
| Age (years) | 32.4 ± 16.1 | 33.8 ± 14.3 | 0.371 |
| Female, n (%) | 29 (85.3) | 37 (82.2) | 0.715 |
| SLE disease duration (months)* | 32.7 ± 31.3 | 27.4 ± 22.6 | 0.143 |
| Rash, n (%) | 21 (61.8) | 23 (51.1) | 0.345 |
| Light sensitivity, n (%) | 10 (29.4) | 14 (31.1) | 0.871 |
| Oral ulcers, n (%) | 13 (38.2) | 17 (37.8) | 0.967 |
| Arthritis, n (%) | 20 (58.8) | 23 (51.1) | 0.496 |
| Serositis, n (%) | 7 (20.6) | 11 (24.4) | 0.686 |
| Kidney damage, n (%) | 29 (85.3) | 29 (64.4) | 0.038 |
| Blood system damage, n (%) | 14 (41.2) | 24 (53.3) | 0.284 |
| CNS involvement, n (%) | 3 (8.80) | 1 (2.20) | 0.42 |
| ANA, n (%) | 34 (100.0) | 44 (97.8) | 1 |
| Anti-dsDNA antibody-positive, n (%) | 22 (64.7) | 24 (53.3) | 0.31 |
| Anti-Sm antibody-positive, n (%) | 12 (35.3) | 18 (40.0) | 0.67 |
| ANA-positive, n (%) | 21 (61.8) | 23 (51.1) | 0.345 |
| Note: ANA: Anti-Nuclear Antibodies; SLE: Systemic Lupus Erythematosus; Sm=Smith; CNS: Central Nervous System. | |||
Table 1. Correlations of different ATG6/Beclin-1 mRNA expression levels with clinical manifestations and laboratory results of SLE.
Discussion
Abnormal immune features of SLE are associated with abnormal activation of T and B lymphocytes, producing a variety of nuclear-targeting autoantibodies and resulting in systemic damages [13,15]. Given the regulatory roles of autophagy in innate and acquired immune systems, eliminating dead cells and intracellular DNA and RNA, and maintaining the generation of T and B lymphocytes, autophagy is currently considered as a key pathogenic contributor to SLE-associated abnormalities [8,12,16-20]. Genomic studies have shown that loci with Single Nucleotide Polymorphisms (SNPs) rs548234, rs6937876, and rs656843 on the autophagy gene locus ATG5 can upregulate ATG5, thus demonstrating a relationship with the occurrence and development of SLE. Further studies have shown that the upregulation of ATG5 is related to the activation of autophagy induced by complement-inactivated serum in patients with SLE [21,22]. Another study reported that the abnormal expressions of the autophagy genes ATG7, LC3, HSPA, UVRAG, and IRGM are related to an imbalance of T-cell functions in patients with SLE [12]. These results suggest that the changes in autophagy gene expressions may play important roles in the pathogenesis or progression of SLE. The formation of autophagosomes is mediated by a group of evolutionarily conserved Autophagy-Related Genes (ATGs) that encode ATG proteins [23], and nearly 36 types of ATG proteins have been found in mammals, among which BECN1 (equivalent to ATG6 in yeast) is a key protein in autophagy initiation; thus, it is considered the gatekeeper of autophagy. As the first known gene directly related to autophagy activation in mammals, BECN1 is a target for inhibiting autophagy activity. After cells receive autophagy-inducing signals, a small membrane-like structure that is similar to a liposome will form in the cytoplasm. This structure is known as a phagophore (equivalent to an autophagic precursor), promoting the activation of downstream autophagic signals and resulting in the interaction of BECN1 with ATG14, VPSl5, and VPS34 to form the PI3KC3 complex. In addition, it recruits FYVE- or PX-containing proteins in the cytoplasm for autophagic membrane formation and to promote other ATG proteins to bind to the phospholipids on autophagic precursors; thus, these autophagic precursors can continue to extend, wrap around cytoplasmic components that need to be reduced (including organelles and other biological macromolecules), and form autophagosomes [24]. Many studies have demonstrated that BECN1 plays crucial roles in inhibiting the occurrence and development of tumors, preventing aging, regulating the immune system, and protecting the nervous system [25-28]. BECN1 biallelic deficiency was shown to result in early placental death in mice, and BECN1 monoallelic deficiency may lead to sporadic tumors and paraplasm of germinal center B cells and easily result in degenerative changes in the nervous system. BECN1 was found to be downregulated in Alzheimer’s disease [29-31]. To investigate the roles of BECN1 expression in the pathogenesis and progression of SLE, we determined and compared BECN1 mRNA expression in the mononuclear cells of the peripheral blood of patients with SLE and of healthy controls. The results demonstrated that the mRNA expression of BECN1 in patients with SLE was significantly reduced (P<001). Further analysis of BECN1 mRNA expression and SLE clinical features revealed that BECN1 mRNA expression level in patients with SLE was significantly negatively correlated with SLEDAI (r=-0.267, P=0.001). Considering that the expression of BECN1 can regulate autophagy, a high expression of BECN1 is consistent with an increased number of autophagosomes. In a previous study, BECN1-deficient rats were found to have significantly reduced number of autophagic vacuoles [24]. Similarly, this study revealed the downregulation of BECN1 mRNA in patients with SLE, indicating that patients with SLE have defects in autophagic function. Another study examined and compared BECN1 mRNA expression in the PBMCs of three patients with SLE and three healthy controls and found that BECN1 mRNA was significantly upregulated in the former [32]. Although the observed BECN1 mRNA upregulation in that study is in contrast to our observation of BECN1 mRNA downregulation in patients with SLE, the previous study may not accurately represent the expression of BECN1 mRNA in patients with SLE because of the limited number of cases. T and B lymphocytes are associated with increased autophagy in SLE, which is also believed to extend the survival period of autoreactive T and B cells to participate in the pathogenesis of SLE [11,19,33]. However, a previous study did not find an increase in autophagy in the T lymphocytes of patients with SLE; conversely, T lymphocytes in SLE were found to resist the stimulation induced by autophagy [34]. In addition, autophagy genes were reported to be downregulated in the macrophages of patients with SLE [34]. Notably, glucocorticoids, the preferred drugs for SLE treatment, may also upregulate autophagy, thus playing a therapeutic role [35,36]. Rapamycin is notable as a specific autophagy activator; in a murine model of lupus (NFBXNZWF1), it was observed that rapamycin can effectively attenuate the reduction of autoantibodies and the deposition of immune complexes and urinary proteins in vasculitis, thereby significantly prolonging the survival period [37]. Clinically, rapamycin has been found to not only suppress SLE disease activity but also significantly reduce the levels of corticosteroids in patients [38]. These results suggest that autophagy deficiency may be related to the pathogenesis or disease activity of SLE, and the results of BECN1 downregulation observed in our study may be associated with existing autophagy deficiency in SLE. Lupus Nephritis (LN) is the most common involvement of a major organ in SLE, and we found that patients with SLE with downregulated BECN1 mRNA had a higher incidence of LN than healthy controls (85.3% vs. 64.4%, P=0.038). The occurrence of LN is thought to be related to anti-dsDNA antibody and Anti-Nuclear Antibody (ANA), and certain evidence has indicated that the autoantibody and autophagy deficiencies in patients with SLE are related to the noneffective removal of apoptotic cells [39]. Therefore, the finding indicating that patients with SLE with downregulated BECN1 mRNA are more susceptible to LN may be attributed to these antibodies. Our results that demonstrated the higher anti-dsDNA antibody levels and ANA-positive rates in patients with SLE with downregulated BECN1 mRNA also support this association.
Overall, our findings are preliminary, and the precise mechanisms leading to the downregulation of BECN1 expression in patients with SLE are not yet well understood. ATG proteins are transcriptionally regulated by several factors such as genes positively or negatively regulating autophagy. An overexpression of genes negatively regulating autophagy has been demonstrated in patients with SLE [34], which might explain the downregulated BECN1 mRNA in our patients. Future studies investigating the role of BECN1 expression in autophagic flux and identifying its downregulation mechanisms may improve our understanding of the biological significance of this gene and its key roles in SLE pathogenesis.
Conflict of Interest
All authors have no conflict of interest regarding this paper.
References
- Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med 2008; 358: 929-939.
- Adinolfi A, Valentini E, Calabresi E, Tesei G, Signorini V. One year in review 2016: systemic lupus erythematosus. Clin Exp Rheumatol 2016; 34: 569-574.
- Squatrito D, Emmi G, Silvestri E, Ciucciarelli L, DElios MM. Pathogenesis and potential therapeutic targets in systemic lupus erythematosus: from bench to bedside. Auto Immun Highlights 2014; 5: 33-45.
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290: 1717-1721.
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90: 1383-1435.
- Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Curr Opin Genet Dev 2014; 26: 16-23.
- Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, Brock R, Driessen C, Rammensee HG, Stevanovic S. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA 2005; 102: 7922-7927.
- Munz C. Enhancing immunity through autophagy. Annu Rev Immunol 2009; 27: 423-449.
- Henckaerts L, Cleynen I, Brinar M, John JM, Van Steen K. Genetic variation in the autophagy gene ULK1 and risk of Crohns disease. Inflamm Bowel Dis 2011; 17: 1392-1397.
- Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb AL. Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy 2009; 5: 152-158.
- Yang Z, Goronzy JJ, Weyand CM. Autophagy in autoimmune disease. J Mol Med (Berl) 2015; 93: 707-717.
- Nagy G, Koncz A, Perl A. T- and B-cell abnormalities in systemic lupus erythematosus. Crit Rev Immunol 2005; 25: 123-140.
- La Cava A. Lupus and T cells. Lupus 2009; 18: 196-201.
- Bennett S, Breit SN. Variables in the isolation and culture of human monocytes that are of particular relevance to studies of HIV. J Leukoc Biol 1994; 56: 236-240.
- Zhou XJ, Zhang H. Autophagy in immunity: implications in etiology of autoimmune/autoinflammatory diseases. Autophagy 2012; 8: 1286-1299.
- Pierdominici M, Vomero M, Barbati C, Colasanti T, Maselli A. Role of autophagy in immunity and autoimmunity, with a special focus on systemic lupus erythematosus. FASEB J 2012; 26: 1400-1412.
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469: 323-335.
- Feng Y, Li B, Li XY, Wu ZB. The role of autophagy in rheumatic disease. Curr Drug Targets 2016.
- Pan Q, Gao C, Chen Y, Feng Y, Liu WJ. Update on the role of autophagy in systemic lupus erythematosus: A novel therapeutic target. Biomed Pharmacother 2015; 71: 190-193.
- Rockel JS, Kapoor M. Autophagy: controlling cell fate in rheumatic diseases. Nat Rev Rheumatol 2016; 12: 517-531.
- Zhou XJ, Lu XL, Lv JC, Yang HZ, Qin LX, Zhao MH, Su Y, Li ZG, Zhang H. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 2011; 70: 1330-1337.
- Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Xu JH, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Li Y, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009; 41: 1234-1237.
- Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jaattela M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell 2007; 25: 193-205.
- Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med 2003; 9: 65-76.
- Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 2010; 17: 268-277.
- Zhu H, He L. Beclin 1 biology and its role in heart disease. Curr Cardiol Rev 2015; 11: 229-237.
- Toton E, Lisiak N, Sawicka P, Rybczynska M. Beclin-1 and its role as a target for anticancer therapy. J Physiol Pharmacol 2014; 65: 459-467.
- OBrien CE, Wyss-Coray T. Sorting through the roles of beclin 1 in microglia and neurodegeneration. J Neuroimmune Pharmacol 2014; 9: 285-292.
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA 2003; 100: 15077-15082.
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999; 59: 59-65.
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA. The autophagy-related protein Beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 2008; 118: 2190-2199.
- Li B, Yue Y, Dong C, Shi Y, Xiong S. Blockade of macrophage autophagy ameliorates activated lymphocytes-derived DNA induced murine lupus possibly via inhibition of proinflammatory cytokine production. Clin Exp Rheumatol 2014; 32: 705-714.
- Zhou XJ, Cheng FJ, Zhang H. Emerging view of autophagy in systemic lupus erythematosus. Int Rev Immunol 2015; 34: 280-292.
- Alessandri C, Barbati C, Vacirca D, Piscopo P, Confaloni A. T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J 2012; 26: 4722-4732.
- Majai G, Kiss E, Tarr T, Zahuczky G, Hartman Z, Szegedi G, Fesus L. Decreased apoptophagocytic gene expression in the macrophages of systemic lupus erythematosus. Lupus 2014; 23: 133-145.
- Harr MW, McColl KS, Zhong F, Molitoris JK, Distelhorst CW. Glucocorticoids downregulate Fyn and inhibit IP (3)-mediated calcium signalling to promote autophagy in T lymphocytes. Autophagy 2010; 6: 912-921.
- Warner LM, Adams LM, Sehgal SN. Rapamycin prolongs survival and arrests pathophysiologic changes in murine systemic lupus erythematosus. Arthritis Rheum 1994; 37: 289-297.
- Fernandez D, Bonilla E, Mirza N, Niland B, Perl A. Rapamycin reduces disease activity and normalizes T cell activation-induced calcium fluxing in patients with systemic lupus erythematosus. Arthritis Rheum 2006; 54: 2983-2988.
- Baumann I, Kolowos W, Voll RE, Manger B, Gaipl U, Neuhuber WL, Kirchner T, Kalden JR, Herrmann M. Impaired uptake of apoptotic cells into tingible body macrophages in germinal centers of patients with systemic lupus erythematosus. Arthritis Rheum 2002; 46: 191-201.