Review Article - Journal of RNA and Genomics (2017) Volume 13, Issue 1
CRISPR/Cas9 genome editing system in the diagnosis and treatment of cancer
Rudrarup Bhattacharjee1, Karishma Das Purkayastha1, Dattatreya Adapa2and Amarendranath Choudhury3,4*
1Department of Life Science and Bioinformatics, Assam University, Silchar-788001, India
2GITAM Institute of Sciences, GITAM University, Visakhapatnam, Andhra Pradesh, India
3Independent Researcher, Kondapur, Hyderabad, Telangana, India
4Alumnus, Department of Life Science and Bioinformatics, Assam University, Silchar, Assam, India
Received: 25 November 2017; Revised: 15 December 2017; Accepted: 18 December 2017; Published: 22 December 2017
© Copyright Amarendranath Choudhury. First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Abstract
While several modes of therapeutic approaches for cancer remained highly debatable, Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (CRISPR/Cas9) tool with enhanced genome editing capabilities has shown promising outcome in recent years. Studies have established the importance of this system in identifying the genes which are critical to a particular type of cancer. According to Cheong and colleagues, CRISPR/ Cas9 system is having profound efficacy in identifying genes associated with oral cancer pathobiology. In another study, 4NQO-induced carcinogenic processing was effectively identified by CRISPR/Cas9 tool. Moreover, in human trial similar results were evident from lung cancer patients. Together, all such novel contributions of CRISPR/Cas9 system have made it a research focus of present time and also established the efficacy of CRISPR/Cas9 system to find out the target for specific set of genes responsible for causing cancer. In Indian perspective, North-East region is having traditional betel-nut chewing habits, which has been reported to be associated with increasing trend of oral carcinogenic processing. Hence, CRISPR/Cas9 system could assist in tracking carcinogenic progression and identifying the expression patterns of genes, which in turn could help to specify potential targets to enhance the global therapeutic possibilities. In the present study, we have discussed all the therapeutic aspects of CRISPR/Cas9 system and presented a rational discussion on the efficacy of CRISPR/Cas9 system in regulation or modification of carcinogenic progression with special emphasis on oral precancerous and cancerous stages.
Keywords
CRISPR/Cas9, Oral cancer, Betel-nut, Gene, Bio-resource
Introduction
Oncogenic cell growth is rapid, heterogeneous and very fast evolving, making it extremely difficult to target specifically. The pathology is generally treated utilizing a mix of chemotherapy, radiation, and surgery. Although the treatments including cytotoxic substances and radiation, which are with blended results and cause adverse side-effects (Urruticoechea et al, 2010). Hence, there is an urgent need of alternative mode of treatment and gene therapy research has got ample of importance in this regards. The point of this emerging field is to alter and convey recombinant DNA for remedial purposes. However, debates and controversies in clinical stages are hindering the eloquent druggability of such methods. It is noteworthy that, during a clinical trial for ornithine transcarbamoylase (OTC) insufficiency, subject Jesse Gelsinger passed on of an insusceptible reaction to the adenoviral vector utilized for conveying the restorative gene (Sibbald, 2001; Jeong et al, 2016). Practically, in the meantime, an exceedingly broadcasted quality treatment trial to treat X-linked Severe combined immune-deficiency (X-SCID) with retrovirus was finished. The treatment appeared like a colossal accomplishment at to start with, however a few patients, later diagnosed with particular diseases because of vector contamination with adjacent protooncogenes and other hereditary issues (Hacein-Bey-Abina et al, 2008). As of now, cancer is the prime focus of quality treatment, making up around 65% of quality treatment and clinical trials starting at 2012 (Wirth et al, 2013). Cancer in Lungs furnishes one of the major reasons for cancer deaths worldwide. Non-small cell lung cancer (NSCLC) is the most widely reported cause of lung cancer, which accounts for 80% of all lung growths and has a 5-year general survival rate of 15%. In the previous five years, genome altering advances utilizing clustered regularly interspersed palindromic repeats (CRISPR) in combination with CRISPR-associated enzyme systems (Cas) brought revolutionary changes in this field (Barrangou and Doudna, 2016). Starting at 2016, two trials for malignancy treatment have been reported in China and the United States, both of which are speculated to use CRISPRCas9 to design and understanding T cells in vitro to eradicate cancer cells (Cyranoski, 2016; Reardon, 2016). Coping with the current scenario, effective gene therapy includes only 5% of all the diagnostic advancements but, considering the worldwide oncogenic growth, addition of CRISPR/Cas9 to this existing therapeutic regime could guarantee better recovery and impressive retardation in carcinogenic. The CRISPR/ Cas9 framework from Streptococcus pyogenes was found as a genome altering instrument for human genome in the year 2012, and current developments in the field of molecular gene editing techniques viz, CRISPR/Cas9 have cleared an approach to create lung malignancy treatment. Epigenetic controllers are frequently disturbed in cancer cells and act as primary mediators which change a normal cell to a malignant one. Cancer-related epigenetic changes or epigenetic factor transformations have a noteworthy impact amid the different strides of carcinogenesis influencing an assortment of disease related qualities along with an extensive variety of destructive phenotypes. Along these lines, epigenetic regulatory enzymes may be hopeful focuses for cancer treatment (Ning et al, 2016; Cui et al, 2015).
CRISPR/Cas9 System in brief
CRISPR/Cas9 is heritable and part of the versatile safe framework in microbes and archaea giving them safeguard against attacking phages and plasmids. CRISPRs and Cas protein locus encodes Cas proteins along with a repeat spacer sequence comprising of mixed indistinguishable repeat sequences along with CRISPR spacer subsystem. A type II CRISPR/Cas9 framework is an adaptive acquired immunity framework, in which CRISPR spacers direct to the target, though Cas enzymes control spacer acquisition and phage defence (Doudna and Charpentier, 2014; Sánchez-Rivera and Jacks, 2015). It was only in 2007, when Barrangou and colleagues discovered that S. thermophilus is able to furnish resistance against a bacteriophage by genome fragment integration into the CRISPR locus in itself (Barrangou et al, 2007). CRISPR mechanisms have three known varieties, of but among them the type II is the one, which is most studied. Target DNA from viruses or plasmids are cut to form tiny fragments and incorporated into a CRISPR locus amidst a series of short repeats (almost 20 bp). Transcription of the loci take place and those are then processed to get small RNAs (i.e, crRNA–CRISPR RNA), which then guide effector endonucleases that focus on incursive DNA based on similarity of sequence (Jinek et al, 2012; Ghorab et al, 2015). Cas9 protein, known also as Csn1 is found to be a major player in many CRISPR mechanisms (especially in type II systems) and it’s one of a kind compared to other CRISPR systems as it requires only one Cas protein i.e, Cas9 for its gene silencing action (Deltcheva et al, 2011). Type II systems employ Cas9 participates to process crRNAs and consequent deletion of the target DNA (Deltcheva et al, 2011; Jinek et al, 2012). For this activity, two nuclease type domain is needed by Cas9, of which one is RuvC-like nuclease domain (amino terminal end) and another is the HNH-like nuclease domain (mid-region of the protein) (Sapranauskas et al, 2011).
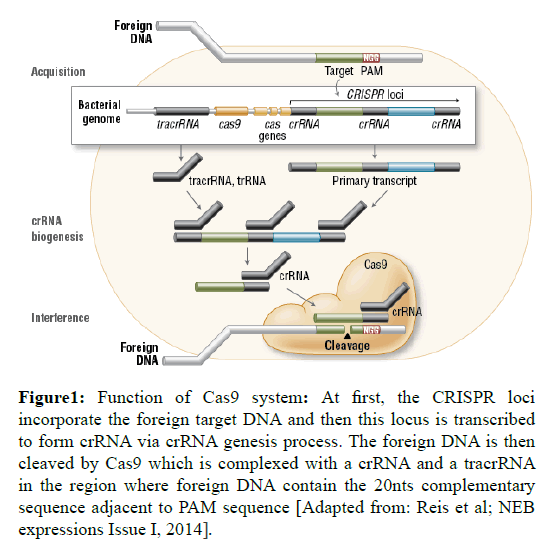
Cas9 is complexed with a crRNA, which functions to degrade invading targetDNA and for that process it also needs a transactivating crRNA (tracrRNA or trRNA) (Jinek et al, 2012). For crRNA maturation, tracrRNA is needed to encode multiple pre-crRNAs and this requires the presence of RNase III and Cas9 (Deltcheva et al, 2011). At the start of target DNA degradation process, the HNH and RuvC-like enzyme domains cut each of DNA strands to produce double-stranded breaks (DSBs) at sites outlined by a 20-nucleotide target sequence associated with crRNA transcript. The HNH domain cuts the complementary strand, while the RuvC domain cleaves the noncomplementary strand (Jinek et al, 2012; Nishimasu et al, 2014). The double-stranded nuclease activity of Cas9 conjointly needs a 2–5 nts long protospacer-associated motif (PAM), which is followed at once 3´- of the crRNA complementary sequence. Fully complementary sequences are also neglected by Cas9-RNA when there is no PAM sequence (Jinek et al, 2012; Nishimasu et al, 2014; Wang et al, 2014) (Figure 1).
Figure 1: Function of Cas9 system: At first, the CRISPR loci incorporate the foreign target DNA and then this locus is transcribed to form crRNA via crRNA genesis process. The foreign DNA is then cleaved by Cas9 which is complexed with a crRNA and a tracrRNA in the region where foreign DNA contain the 20nts complementary sequence adjacent to PAM sequence [Adapted from: Reis et al; NEB expressions Issue I, 2014].
CRISPR/Cas9 as a genome editing tool
In the previous five years, genome altering innovations by utilizing CRISPR in combination with Cas have redefined the therapeutic possibilities in related fields of studies (Sánchez- Rivera and Jacks, 2015). The innovation has turned out to be so flexible and open that it has been embraced in scholastic labs worldwide and has been highlighted in more than 5000 distributions on PubMed since 2013. The organization of CRISPR/Cas9 advances has even offered ascend to human germ-line altering discussions (Doudna and Charpentier, 2014). Starting at 2016, two trials for tumor treatment have been reported in China and the United States, both of which will use CRISPR/Cas9 to create understanding the T cells in vitro to wreck disease cells (Cyranoski, 2016; Lindsay-Mosher and Su, 2016). In spite of the fact that, quality treatment accounts 5% of interventional cancer studies worldwide right now, this improvement has impressive promise for future. Usage of this innovation needs a Cas nuclease most commonly from the bacterium Streptococcus pyogenes, to be communicated inside the cell of interest. During experimentation, Cas9 requires a guide RNA (gRNA) composed of a systematic arrangement for Cas9 system and a spacer region that characterizes the target locale (Hsu et al, 2013). Infusing bare plasmids encoding Cas9 and a gRNA into the circulatory system brings about low levels of gene editing in mice (Yu et al, 2013). New conveyance strategies for CRISPR-Cas9 should have effectiveness without toxicity, be able to bypass host defence system, and should be specific to target cancer cells and not normal ones. The in vitro and pre-clinical advances in creating viral and non-viral vectors for CRISPR/Cas9 conveyance made within couple of years of time, and discussion about their suggestive use for cancer treatment was done by Mosher et al,. (Lindsay-Mosher and Su, 2016). The most prominent instruments for conveyance of CRISPR/Cas9-intervened gene therapy today are viral vectors, which made up about 66% of gene therapy trials currently (Sibbald, 2001) . The challenge of utilizing viral vectors is to guarantee that they are particular in their objective and in their tropism (i.e, likeliness for a particular cell) (Lindsay-Mosher and Su, 2016). To exacerbate the specificity issue, CRISPR/ Cas9 quality altering is likewise subject to off-target impacts, which have been widely examined. Adeno-Associated Virus (AAVs), adenoviral vectors (AdVs), and retroviruses have contained the majority of gene therapy trials to date. AdVs hold the profit of conveying bigger constructs than AAVs, with a more elevated amount of related protein expression, and have been utilized to convey Cas9 with coming about targeting on effectiveness to rates accomplished with Translation Activator- Like Effector Nucleases (TALENs) (Wirth et al, 2013). Retroviruses change over their viral RNA genome to DNA through reverse transcriptase and incorporate into the genome. As suggested from X-SCID trial and other gene therapy trials, coordinating infections can be perilous because of associated insertional oncogenesis threat (Kawamura et al, 2015). Ironically, in the OTC trial, adenoviruses were deadly because of their high immunogenicity. Another approach to lessen the dangers of insertional oncogenesis and immunogenicity is to utilize AAVs set up of AdVs. AAVs have a nearly low occurrence of reconciliation in the genome and hold on as episomes in primates (Urruticoechea et al, 2010). Although, vast majority of the populace carries antibodies for a few strains of (AAV-1 and AAV-2), prevalence of antibodies for different strains, for example, AAV-5, AAV-6, AAV-8 and AAV-9 is much lower and accordingly these alternatives might be reasonable for gene therapy (Kawamura et al, 2015; Lindsay- Mosher and Su, 2016; Lin et al, 2015). Consolidating AAVs with CRISPR/Cas9 guarantees an answer which permits differing and enduring quality altering with negligible immunogenic response. AAVs are the smallest viral vectors, with a genome size of around 4.7 kB. In any case, Cas9 can be part and afterward practically reconstituted by, including autoprocessing excisable protein domains (Yu et al, 2013). Moreover, recent studies utilizing AAV-9 to convey split Cas9 in mouse models did not suggest invulnerable reaction to the viral vector itself, even when antibodies were made against the Cas9 protein ( Wang et al, 2017; Ye et al, 2015). The contrasting options to AAVs, AdV’s and retroviruses are rising. The issues of safety, immunogenicity, and payload estimate in viral vectors have incited investigate into elective methods of DNA conveyance in vivo. Except viral vectors, engineered vectors don’t contain immunogenic pathogen-related molecules, and patients are therefore unlikely to have pre-existing immunity (Sachdeva et al, 2015). The important inconvenience of engineered vectors is their low gene transfer efficiency contrasted with viral vectors, which restrains their utility for both systemic and intra tumoral (Jobin Christ et al, 2015) conveyance. Advances in the plan of lipid and polymeric vectors have expanded the practicality of conveying DNA and RNA for treatment in vivo; in any case, conveyance of CRISPR/Cas9 presents a signature trouble of its own. Though, manufactured lipid or polymer nanoparticles can undoubtedly be made to convey a payload significantly bigger than 4.2 kb, which is the size of Cas9 nanoparticles should contain a small volume as the final output is conveyed fundamentally through the endothelial gaps in veins (Kawamura et al, 2015; Sun et al, 2015, Sun et al, 2016). This implies the grouping of DNA within the nanoparticles must be high keeping in mind the end goal to convey CRISPR/Cas9 intravenously. Likewise, as with viral vectors, conveyance of DNA encoding CRISPR/Cas9 risks irregular joining into the genome. To address these issues, further studies utilized non-virus based vectors to convey Cas9 protein instead of using DNA in vivo, because former is unlikely to cause insertional oncogenesis by means of genome integration. Conveyance of Cas9 protein in mouse models utilizing lipid and polymeric nanoparticles has demonstrated particularly encouraging. Cas9 in recent studies was effectively conveyed to mammalian cells utilizing cationic lipid nanoparticles (liposomes) (Sun et al, 2015; Doganlar et al, 2016). However, Cas9 itself isn’t adequately anionic for conveyance with cationic lipids, complexing Cas9 with a gRNA in-wrinkled the negative charge on the protein. Transfection of human cells in vitro demonstrated that, protein conveyance was somewhat more productive than plasmid conveyance, and brought about a ten times decrease in nonspecific genome editing. Another study showed that infusion of the Cas9-gRNA-lipid complex into the mouse internal ear brought about 20% transfection of mouse inward ear cells, with no recognizable toxicity. A subsequent report could enhance the transfection proficiency by using bio reducible lipids, which degrade in the reductive condition of the cell, enabling the payload to be discharged after endosomal escape (Wang et al, 2017; Wang et al, 2014; Wang et al, 2016). Utilizing this strategy, the transfection effectiveness of human cells with CRISPR/Cas9 in vitro was expanded from roughly 40% to more than 70%. Despite in vivo altering with CRISPR/ Cas9 was not endeavoured, the nanoparticles of lipid were utilized to convey an alternate genome altering protein, Crerecombinase, to mouse cerebrum cells close to the site of infusion. Nanoparticles made of palindromic DNA, known as ‘nanoclews’, have additionally been utilized to convey CRISPR/ Cas9 to tumor cells in vivo. The nanoclews were complexed with the cationic polymer PEI to counterbalance the negative charge of the DNA spine (Sun et al, 2015; Tian et al, 2016). These outcomes demonstrate that lipid nanoparticles can be utilized to convey CRISPR/Cas9 foundationally; however, these nanoparticles were not tissue specific and consequently transfected a few non-target tissues with moderately low efficiency. To target growth fundamentally, engineered nanoparticles should be intended to explicitly transfect malignancy cells. Nanoparticles being produced by various materials have only one major goal to accomplish this objective of such transfer. An assortment of engineered vectors including liposomes, bio reducible lipids, nanoclews indicate guarantee for conveyance of CRISPR/Cas9 in vitro. By and large, examines using engineered vectors to treat disease utilizing CRISPR/Cas9 in vivo should be completed before the capability of non-viral vectors can be figured it out (Sun et al, 2015) (Table 1).
| CRISPR Associated Functional Units | Function | References |
|---|---|---|
| Cas:CRISPR-assocaited genes | Utilized by S. pyogenes primarily for memory and also to save itself from invading viruses | (Heler et al, 2015) |
| Cas9, Csn1: A CRISPR associated gene product i.e., protein having two nuclease domains, programmed by small RNAs to cleave DNA | Guide RNA mediated target sequence cleavage | (Jinek et al, 2012) |
| crRNA: CRISPR associated RNA | They form effector complex and guide the nuclease system to the invading genetic material to degrade it | (Deltcheva et al, 2011) |
| dCAS9: Nuclease deficient Cas9 Protein/catalytically dead Cas9 | Lost double stranded DNA break ability but retain single stranded target identification ability | (Jinek et al, 2012) |
| DSB: Double stranded break | Both the strands of DNA are broken , primarily happens due to radiation but also can occour due to DNA lesion or oncogenetic progression | (Acharya, 1972) |
| gRNA: Guide RNA | These are crucial for Cas function as they assist Cas to cut at specific genomic location | (Brouns et al, 1993) |
| HDR: Homology-Directed Repair | Double stranded break repair by homologous recombination, only functional when there is homologue of DNA present, mostly in s phase or G2 phase in nucleus | (Bolderson et al, 2009; Pardo et al, 2009) |
| HNH: an endonuclease domain named for having characteristic histidine and asparagine residues | Present in most enzymes and serve as catalytic centre for endonucleases, very useful due to its flexibility | (Yusufzai and Kadonaga, 2010) |
| Indel: Insertion and/or deletion | insertionordeletionofbasesin genome which consist variations from 1 to 10 000 basepair | (Gregory, 2004) |
| NHEJ: Non Homlogous End Joining | Non homologous end joining repair using microhomologies | (Moore and Haber, 1996) |
| PAM: Protospcer-Adjacent Motif | A sequence of 2 to 6 base pair present adjacent to crispr target sequence. It is mandatory for successful Cas9 function | (Shah et al, 2013) |
| RuvC: an E. coli endonuclease involved in DNA repair | Holiday junction resolving enzyme called resolvase, necessary for DNA repair | (West, 2003) |
| sgRNA: Single guide RNA | Single guide RNA for specific target sequence binding | (Brouns et al, 1993) |
| TALEN: Transcription-Activator Like Efector Nuclease | These are restriction enzymesthat are used to cut specific sequences of DNA. They utilize TAL effector DNA-binding domainto a DNA cleavage domain | (Boch, 2011) |
| ZFN: Zinc Finger Nuclease | Artificial restriction enzymes which are generated by fusion of aDNA cleavage domain to Zinc Finger DNA binding domain | (Ramirez et al, 2008) |
| tracrRNA, trRNA: Trans activating crRNA | Trans activating crRNA needed for efficient CRISPR/Cas genome editing | (Deltcheva et al, 2011) |
Table 1: Some Common Terms associated with CRISPR/Cas9 editing system
Recent trials using CRISPR-Cas9 system
The RNA-guided nuclease CRISPR/Cas9 has recently emerged as an efficient tool (Jinek et al, 2012) and hence led to great number of articles utilizing this process for genome editing in diverse array of organisms including bacteria (Jiang et al, 2013), yeast (DiCarlo et al, 2013), worms (Friedland et al, 2013), fruit flies (Gratz et al, 2014), zebrafish (Hwang et al, 2013), mammalian cell lines (Cong et al, 2013), mice (Wang et al, 2014), plants (Nekrasov et al, 2013), and food crops such as rice and wheat (Shan et al, 2013). Additionally, Cas 9 has the potential to specifically insert regulatory sequences which ultimately helps in gene expression regulation (Gilbert et al, 2014; Qi et al, 2013). Excellent efficacy in perturbation and editing of genomes with precision has granted this system enormous experimental manipulation ability. In C. elegans, transposon guided genome editing was most common which then produce double stranded break upon excision. When a template of repair is absent, excision of an endogenous Tc1 transposon can produce gene mutations or deletions (Kawamura et al, 2015). Likewise, generation of transgenic lines with extrachromosomal arrays which are semi-stable and contain multiple copies of injected DNA, are relatively simple (Mello et al, 1991) and they provide repairing templates for double stranded break when the Tc1 transposon gets excised (Plasterk, 1992). One pioneering example of endogenous gene editing, including tagging a gene with GFP, was based on Tc1 excision (Barrett et al, 2004). However, the process was with lot of disadvantages, one of which was Tc1 is activity only in mutator strains. The strains containing several classes of active transposons and having multiple transposition events ultimately generated high mutational load (Ruaud and Bessereau, 2006). Later, the Mos1 transposon from Drosophila was used to produce improved gene-editing methods (Bessereau et al, 2001). To generate specific deletions and insertions, single Mos1 transposon excision was carried out (Boulin and Bessereau, 2007) and also affinity tags were inserted including GFP via extra-chromosomal plasmids insert without inducing breaks throughout the genome. The effort of a consortium of European labs which generate a collection of 13,000 strains carrying Mos1 elements enhanced the usefulness of this system to many folds (Bazopoulou and Tavernarakis, 2009; Vallin et al, 2012). Insertion of single copies of transgenes into specific genomic locations were then possible utilizing these resources and they also facilitated targeted deletions in large quantities (>25 kb) using positive and negative selection markers (Frøkjær-Jensen, 2013; Frøkjær-Jensen et al, 2008; Frøkjær-Jensen et al, 2012). Genetic manipulation in C. elegans was met with one challenge and that was the inability to generate sustained expression in the germ line from extra chromosomal transgenes (Kelly et al, 1997). However, they could attain success because of transient expression of injected transgenes in the germ-line (Kelly et al, 1997), and this mobilized Mos1 elements effectively, using strong ubiquitous eft-3 promoter (Frøkjær-Jensen et al, 2008, 2012). The genome modification ability is not limited to locations near endogenous or exogenous transposons because of fast improvements in engineered DNA nucleases. The first synthetic technique, that generated double stranded break at targeted genomic regions, was based on chimeric proteins consisting of a DNA binding domain (a zinc finger) and a nonspecific nuclease (FokI) (Bessereau et al, 2001). These zinc finger nucleases (ZFNs) were engineered to work in pairs to increase break specificity by binding of two chimeric proteins at nearby DNA sequences for nuclease activity (Smith et al, 2000). Although, the DNA sequence recognition code of zinc fingers is complicated, but it must be selected experimentally to ensure high efficiency of each ZFN in work (Maeder et al, 2008).
A new era of oral cancer genetics
Most of the oral cancer patients only know their condition when the disease has almost spread or at least at an advanced stage. The common therapies are surgical and chemo-therapy/radiotherapy, which are accompanied by severe side effects. So, it became necessary to find target based therapies and earlier detections to prevent or at least cure the disease at its very onset. In this light, Cheong and colleagues from University of Malaya has come up with new genome based identification for oral cancer and they even developed an oral cancer vaccine, which is soon going to be in human clinical trials. Collaborating with Dr McDermott and Well-come Trust, this group is now to use CRISPR/Cas9 system to specifically target key genes which express in case of oral cancer and they try to silence them by knock out. Cheong and colleagues have developed 16 different cell lines for carrying out this experiment and more are still being tested. They are also trying to deal with the multidrug resistance of cancer cells and its underlying genetics. These experiments show quite good results and paves the way to further advanced genetic research using CRISPR genome editing to finally get an answer towards oral oncology genetics (Ruaud and Bessereau, 2006).
Future prospects of the CRISPR technology
Although varied drugs and therapies have been developed for lung cancer treatment, over the past 5 years overall survival rates have not improved much. Cas9 can target any gene in a tissue-selective manner (tissue-specific promoters in plasmid or viral construct) inhibiting, repressing, activating, translocating, inverting or duplicating them, and hence CRISPR/Cas9 may serve as a boon for lung cancer therapeutics. Overexpressed epidermal growth factor receptor has been reported to be repressed by the use of this technique, which might result in effective study of tyrosine kinase inhibitor or Alk-activated pathways, which otherwise was in infancy. RNAi mediated gene therapy identifies K-RAS as one of the primary targets; CRISPRi is a perfect candidate to overtake RNAi technology because the former has increased number of advantages, of which no competition with endogenous machinery such as miRNA is a noteworthy one (Jiang et al, 2013). In addition, CRISPR/Cas9 activity is at DNA level, and hence it helps targeting transcripts like noncoding RNAs, miRNAs, antisense transcripts, nuclearlocalized RNAs and polymerase III transcripts. Due to diverse and manipulative nature, it also possesses a much larger targetable sequence space. All the recent advances along with the upcoming ones would help in developing and optimizing Cas9-based systems for genomic and epigenomic prints that will propel the technology toward therapeutic applications, widening the path for treating a vast array of human diseases (Biagioni et al, 2017). Future advancement of these techniques could give rise to a completely new pharmacological treatment class that might contribute to cure by altering disease-associated genomics and epigenomic signatures (Biagioni et al, 2017; Jiang et al, 2013; Reardon, 2016; Shan et al, 2013).
Conclusion
The oral cancer incidents are a persistent threat to human wellbeing and most of it either get diagnosed late or ill-treated. Operational therapies although provide relief but at the cost of various systemic damage to the patient in question. In this evergrowing risk of developing oral carcinoma, the CRISPR/Cas9 genome editing systems have proven to be a new milestone. Early detection of the disease with early marker identification and selective gene silencing using the CRISPR system may help tackle the disease more efficiently than ever. This toll can not only be used for prophylactic measure but knowing the exact genetic cascade of oral cancer progression, the key genes helping the cancer cell can also be silenced/modified. The CRISPR system is relatively new and further more precise investigations are warranted to establish it as a potent therapeutic tool for cancer diagnosis/treatment. The modification of this system along with specific site directed binding and action could lead to a very fruitful result in the field of cancer therapy. The potential of this tool is huge and selective optimizations and conjugation with existing therapies could provide a more secure future for noninvasive cancer therapeutics.
References
- Acharya PV. 1972. The isolation and partial characterization of age-correlated oligo-deoxyribo-ribonucleotides with covalently linked aspartyl-glutamyl polypeptides. Johns Hopkins Med J Suppl, 1, 254-260.
- Barrangou R and Doudna JA. 2016. Applications of CRISPR technologies in research and beyond. Nat Biotechnol, 34, 933-941.
- Barrangou R, Fremaux C, Deveau H, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science, 315, 1709-1712.
- Barrett LF, Quigley KS, Bliss-Moreau E, et al. 2004. Interoceptive sensitivity and self-reports of emotional experience. J Pers Soc Psychol, 87, 684–697.
- Bazopoulou D and Tavernarakis N. 2009. The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans. Genetica, 137, 39–46.
- Bessereau JL, Wright A, Williams DC, et al. 2001. Mobilization of a drosophila transposon in the Caenorhabditis elegans germ line. Nature, 413, 70-74.
- Biagioni A, Chillà A, Andreucci E, et al. 2017. Type II CRISPR/ Cas9 approach in the oncological therapy. J Exp Clin Cancer Res, 36, 80.
- Boch J. 2011. TALEs of genome targeting. Nat Biotechnol, 29, 135-136.
- Bolderson E, Richard DJ, Zhou BBS, et al. 2009. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res, 15, 6314- 6320.
- Boulin T and Bessereau JL. 2007. Mos1-mediated insertional mutagenesis in Caenorhabditis elegans. Nat Protoc, 2, 1276– 1287.
- Brouns SJJ, Jore MM, Lundgren M, et al. 2008. Small Crispr Rnas Guide Antiviral Defense in Prokaryotes. Science, 321, 960–964.
- Cong L, Ran FA, Cox D, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819– 823.
- Cui YG, Xue F, Lin F, et al. 2015. Extraction process of ginger Pinellia and its anti-proliferative and proapoptotic activities on human gastric cancer SGC7901 cells. Biomed Res- India, 27.
- Cyranoski D. 2016. CRISPR gene editing tested in a person. Nature, 539, 479.
- Deltcheva E, Chylinski K, Sharma CM, et al. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature, 471, 602–607.
- DiCarlo JE, Norville JE, Mali P, et al. 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res, 41, 4336-4343.
- Doganlar O and Doganlar ZB. 2016. Evaluation of the selective anticancer potential and the genetic mechanisms of the induction of apoptosis by walnut milk in human breast and prostate cancer cells. Biomed Res- India, 27, 268–278.
- Doudna JA and Charpentier E. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science, 346, 1258096–1258096.
- Friedland AE, Tzur YB, Esvelt KM, et al. 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods, 10, 741-743.
- Frøkjær-Jensen C. 2013. Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9. Genetics, 195, 635-642.
- Frøkjær-Jensen C, Davis MW, Ailion M, et al. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods, 9, 117-118.
- Frøkjær-Jensen C, Wayne DM, Hopkins CE, et al. 2008. Singlecopy insertion of transgenes in Caenorhabditis elegans. Nat Genet, 40, 1375-1383.
- Ghorab MM and Alsaid MS. 2015. Anticancer activity of some novel thieno [2, 3-d] pyrimidine derivatives. Biomed Res- India, 27.
- Gilbert LA, Horlbeck MA, Adamson B, et al. 2014. Genomescale CRISPR-mediated control of gene repression and activation. Cell, 159, 647-661.
- Gratz SJ, Ukken FP, Rubinstein CD, et al. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in drosophila. Genetics, 196, 961-971.
- Gregory TR. 2004. Insertion-deletion biases and the evolution of genome size. Gene, 324, 15-34.
- Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest, 118, 3132-3142.
- Heler R, Samai P, Modell JW, et al. 2015. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature, 519, 199-202.
- Hsu PD, Scott DA, Weinstein JA, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol, 31, 827-832.
- Hwang WY, Fu Y, Reyon D, et al. 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol, 31, 227-229.
- Jeong SH, Jiang Y, Guo H, et al. 2016. Anti-inflammatory and anticancer effects of methanol, ethanol and water extracts of asiasarum heterotropoide. Biomed Res- India, 27, 103–109.
- Jiang W, Bikard D, Cox D, et al. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol, 31, 233-239.
- Jinek M, Chylinski K, Fonfara I, et al. 2012. A programmable Dual-RNA-Guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816-821.
- Jobin Christ MC and Subramanian R. 2015. Clown fish queuing and switching optimization algorithm for brain tumor segmentation. Biomed Res- India, 27.
- Kawamura N, Nimura K, Nagano H, et al. 2015. CRISPR/ Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget, 6, 22361-22374.
- Kelly WG, Xu S, Montgomery MK, et al. 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics, 146, 227-238.
- Lindsay-Mosher N and Su C. 2016. Cancer gene therapy: innovations in therapeutic delivery of CRISPR-Cas9. Drug Discov Today Dis Models, 21, 17-21.
- Lin J, Yao HJ and Li RY. 2016. Bakuchiol inhibits cell proliferation and induces apoptosis and cell cycle arrest in SGC-7901 human gastric cancer cells. J BUON, 21, 889-894.
- Maeder ML, Thibodeau-Beganny S, Osiak A, et al. 2008. Rapid “Open-Source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Molecular Cell, 31, 294-301.
- Mello CC, Kramer JM, Stinchcomb D, et al. 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J, 10, 3959-3970.
- Moore JK and Haber JE. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol, 16, 2164-2173.
- Nekrasov V, Staskawicz B, Weigel D, et al. 2013. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol, 31, 691- 693.
- Ning B, Li W, Zhao W, et al. 2016. Targeting epigenetic regulations in cancer. Acta Biochim Biophys Sin Shanghai, 48, 97-109.
- Nishimasu H, Ran FA, Hsu PD, et al. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell, 156, 935-949.
- Pardo B, Gómez-González B and Aguilera A. 2009. DNA repair in mammalian cells. Cell Mol Life Sci, 66, 1039-1056. Plasterk RHA. 1992. Genes and genomes: Reverse genetics of Caenorhabditis elegans. BioEssays, 14, 629-633.
- Qi LS, Larson MH, Gilbert LA, et al. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152, 1173-1183.
- Ramirez CL, Foley JE, Wright DA, et al. 2008. Unexpected failure rates for modular assembly of engineered zinc fingers. Nature Methods, 5, 374–375.
- Reardon S. 2016. First CRISPR clinical trial gets green light from US panel. Nature.
- Ruaud AF and Bessereau JL. 2006. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development, 133, 2211-2222.
- Sachdeva M, Sachdeva N, Pal M, et al. 2015. CRISPR/Cas9: Molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Ther, 22, 509-517.
- Sánchez-Rivera FJ and Jacks T. 2015. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer, 15, 387-395.
- Sapranauskas R, Gasiunas G, Fremaux C, et al. 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res, 39, 9275- 9282.
- Shah SA, Erdmann S, Mojica FJM, et al. 2013. Protospacer recognition motifs. RNA Biology, 10, 891-899. Shan Q, Wang Y, Li J, et al. 2013. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol, 31, 686-688.
- Sibbald B. 2001. Death but one unintended consequence of gene-therapy trial. CMAJ, 164, 1612.
- Smith T, Groen AD and Wynn JW. 2000. Randomized trial of intensive early intervention for children with pervasive developmental disorder. Am J Ment Retard, 105, 269.
- Sun W, Ji W, Hall JM, et al. 2015. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl, 54, 12029-12033. Sun DY, Yu H, Qiu X, et al. 2016. Relationships between CD44, hyaluronic acid expression and lymphatic metastasis and radiosensitivity of nasopharyngeal carcinoma. Biomed Res- India, 27.
- Tian H, Liu L, Yang T, et al. 2016. Gentiopicroside inhibits cancer cell growth in OVCAR-3 ovary cancer cells through the mediation of apoptosis, loss of mitochondrial transmembrane potential and NF-kB signalling pathway. Biomed Res- India, 27, 413-418.
- Urruticoechea A, Alemany R, Balart J, et al. 2010. Recent advances in cancer therapy: an overview. Curr Pharm Des, 16, 3-10.
- Vallin E, Gallagher J, Granger L, et al. 2012. A genome-wide collection of Mos1 transposon insertion mutants for the C. elegans research community. PLoS One, 7, e30482.
- Wang H, Xue Y, Di W, et al. 2016. Preparation process of paclitaxel liposomes and its anti-proliferative effect on human gastric cancer MGC803 cells. Biomed Res- India, 27, 360- 363.
- Wang JZ, Wu P, Shi ZM, et al. 2017. The AAV-mediated and RNA-guided CRISPR/Cas9 system for gene therapy of DMD and BMD. Brain Dev, 39, 547-556.
- Wang T, Wei JJ, Sabatini DM, et al. 2014. Genetic Screens in human cells using the CRISPR-Cas9 system. Science, 343, 80-84.
- West SC. 2003. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol, 4, 435-445. Wirth T, Parker N and Ylä-Herttuala S. 2013. History of gene therapy. Gene, 525, 162-169.
- Ye H, Ye G, Jiang J, et al. 2015. Anti-proliferative effect of allicin on human hepatoma HepG2 cells. Biomed Res- India, 27.
- Yu Z, Ren M, Wang Z, et al. 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in drosophila. Genetics, 195, 289-291.
- Yusufzai T and Kadonaga JT. 2010. Annealing helicase 2 AH2, a DNA-rewinding motor with an HNH motif. Proc Natl Acad Sci U S A, 107, 20970-20973.