Research Article - Biomedical Research (2016) Health Science and Bio Convergence Technology: Edition-I
Computerized diabetic patients fundus image screening for lesion regions detection and grading
Ratna Bhargavi V1*, Ranjan K. Senapati1, Gandharba Swain2 and P.M.K Prasad31Department of Electronics and Communication Engineering, K L University, Vaddeswaram, Guntur, Andhra Pradesh, India
2Department of Computer Science and Engineering, K L University, Vaddeswaram, Guntur, Andhra Pradesh, India
3Department of Electronics and Communication Engineering, GMR Institute of Technology, Rajam Srikakulum, Andhra Pradesh, India
- *Corresponding Author:
- Ratna Bhargavi V
Department of Electronics and Communication Engineering
K L University
Andhra Pradesh
India
Accepted date: October 15, 2016
Abstract
Damaging of anatomical structures like vessels in eyes, kidneys, heart and nervous systems occurs in people suffering with diabetes. Diabetic Retinopathy (DR) is an important hurdle in diabetic people and it is main root cause of lesion formation in retina. Bright Lesions (BLs) are preliminary clinical sign of DR. Early BLs detection can help avoiding blindness. The severity can be recognized based on increasing number of BLs formation in the color fundus image. Manually diagnosing large amounts of images is time consuming. So a computer-assisted DR evaluation and BLs detection system is proposed. In this paper initially curvelet enhancement is done for unwanted patches removal. For BLs detection, the Optic Disk (OD) and vessel structures are segmented and eliminated by thresholding techniques. Because OD and vessels are obstacles for exudates exact recognition and DR severity grading. Few images were obtained from ABC imaging centre located in Vijayawada, Andhra Pradesh for testing of the proposed. Publicly available databases are also used for DR severity testing. The Support Vector Machine classifier (SVM) is a supervised learning technique separated fundus images in various levels of DR based on extracted feature set. The proposed system obtained the better results compared to the existing techniques in terms of statistical measures sensitivity, specificity and accuracy.
Keywords
Diabetic retinopathy, Curvelet, Bright lesions, Feature extraction, Classification, Segmentation, Support vector machine classifier.
Introduction
Diabetes is one of the prevalent, progressing diseases in all over the world [1,2]. Commonly, people with diabetes are having DR causes retinal damage. If it is recognized in the early stage, vision loss can be avoided by having laser treatment or other therapeutic techniques. DR causes severe damage of retina blood vessels. The damaged vessels discharge blood and other fluids. Then it causes swelling of retina and lesion formation and it is known as Non-Proliferative DR (NPDR). There is another type of DR occurs that is proliferative DR (PDR). PDR occurs due to damage of blood vessels, blood flow prevention and new vessels formation. These new vessels do not supply proper blood to retina and may cause retinal detachment. In this paper we are recognizing bright lesions or exudates are the pathologies which appear bright yellow or white color with varying sizes and shapes. So the yellow color patches should be identified in the early stage to prevent progression to blindness [3]. Various levels of NPDR severity grading occurs. These are: No DR, Mild Non-Proliferative DR (NPDR), Moderate NPDR, and Severe NPDR [4]. For the treatment of the disease, the DR severity grading determination is necessary and it is shown in Table 1 [5].
| DR stage | Description |
|---|---|
| Grade 0 (no DR) | Pathologies are nil. No red lesions and BLs. |
| Grade 1 (mild) | It is preliminary stage of DR and numerous lesions start to develop. But vision is normal except in some cases. |
| Grade 2 (moderate) | There is no greater severity of red lesions but BLs will be formed from leakage of damaged vessels. |
| Grade 3 (severe) | Severe damage to vessel structures. Large number of red and BLs formed in four quadrants of fundus image. Automated screening of retina for the patients with diabetes in early stage is important. This can save diabetic patient’s vision. So developing of Computer aided detection system for DR screening will yield less time consumption for huge number of patients. |
Table 1. DR Grading stages.
For automatic DR grading, various techniques are implemented by many researchers. Successive clutter rejection with feature extraction is implemented in [6]. It gives true lesions recognition and rejection of false lesion regions. Quellec et al. [7] proposed wavelet transform based matching using a Gaussian template. A multilayer neural network employed to classify the detection of exudates or BLs [8]. They used Genetic algorithm for features selection from large number of features. The obtained sensitivity and specificity are 96%, 94.6% respectively. Hatanaka et al. [9] proposed hue, saturation and mahalanobis distance for the detection of lesions. They tested the technique on 125 images. Combination of fuzzy neural network classifier utilized for dark and bright lesion detection [10]. For exudates regions detection morphological closing operation on luminance channel and watershed transform is applied in [11]. In [12] higher order spectra based features are proposed. SVM is used for classification of lesions, based on mentioned features. They graded fundus images in various stages with average accuracy of 82%. An automated detection of exudates and other lesions was presented in [13]. The reported accuracies for exudates and other lesions are 82.6% and 88.3% respectively. Grading of various stages of DR is reported based on feed-forward neural network in [14]. The used features were area and perimeter of Vasculature structure of color fundus images. Vallabha et al. [15] used global image feature extraction to categorize mild NPDR and severe NPDR. They identified vascular abnormalities based on scale and orientation selective Gabor filters banks. Tan et al. [16] proposed macula detection based on low pixel intensity. Based on ARGALI (An automatic cup-to-disc ratio measurement system for glaucoma analysis using level set image processing) method height of OD is computed to have the ROI (region of interest) of macular region. The algorithm obtained the AUC as 0.988. In [17] exudates are detected by removing the OD and vessel parts. Exudates in various stages of DR are identified. They implemented a sequence of image processing techniques for lesion detection. Wang et al. [18] used adjustment of brightness in image and window based verification method for exudates identification. They were achieved 100% sensitivity and specificity 70%. Osareh et al. [19] used different types of machine learning techniques. The used neural network given better results compared to SVM classifier. The obtained sensitivity and specificity are 93%, and 94.1% respectively. For OD localization, morphological filters and watershed transform implemented [20]. The exudates are detected based on gray level variation and the boundaries are identified by morphological techniques. Results shows sensitivity and specificity are 92.8%, and 92.4%. Fuzzy C-means clustering and SVM are implemented in [21] for BLs detection and classification. The reported sensitivity and specificity are 97%, 96%. Multi-scale morphological techniques are implemented to get the candidate exudates [22]. They used 13219 images for testing and obtained sensitivity and specificity are 95% and 84.6% respectively. The healthy and diseased DR retina was categorized based on image processing techniques and multilayer perceptron neural network [23]. They reported sensitivity 80.21% and specificity 70.66%. Usher et al. implemented a support system for DR screening [24]. Their system got the sensitivity of 95.1% and specificity of 46.3%.
Philip et al. designed an automated system for grading disease or no disease within the screening system [25]. The system achieved 90.5% sensitivity and 67.4% specificity. Abramoff et al. [26] used the existing published methods. They tested the system with large amount of people for screening. The achieved sensitivity and specificity are 84%, 64% for their methodology (Table 1).
Automated screening of retina for the patients with diabetes in early stage is important. This can save diabetic patient’s vision. So developing of Computer aided detection system for DR screening will yield less time consumption for huge number of patients.
Therefore, the objective of the proposed work is to design an automated detection system that can be used for lesion detection and DR grading for various stages (e.g., Grades 0-3) accurately. Figure 1 shows the healthy (Grade 0) retinal image for illustration. The proposed system consists of preprocessing, localization of OD, and elimination of both OD as well as vessel structures. Exudates detection and DR grading using SVM is carried out. The elimination of OD and vessel parts yields better exudates detection and false positive reduction.
The rest of the paper description is as follows: Section 2 describes pre-processing, detection and elimination of Optic disk, identification of lesion parts, feature extraction and then classification. The analysis of obtained results is presented in Section 3. At lastly, conclusions and future implementations are presented in Section 4.
Proposed Method
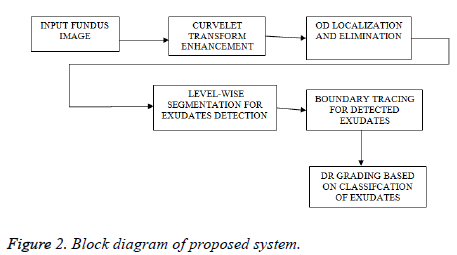
The proposed method comprises various stages for DR grading. Initially, curvelets are used to reduce noise in fundus images. Then the identification and elimination of OD is done using Hough-man transform. After the elimination of anatomical structures based on thresholding, the pathological parts have been identified. Finally DR grading is done based on classification. Figure 2 showing the block diagram of proposed technique and the following sections will give detailed explanation about each method.
Image pre-processing
The pre-processing for color fundus images reduces or removes the effects of noises and unwanted patches visible like lesions. For images denoising in multi-scale regions, Curvelet transform is good for capturing edge discontinuities in all coefficients. In this system, curvelet transform via wrapping [27] is implemented in color fundus image for its fast computation time.
The color fundus images with various sizes and shapes appeared bright lesions having resolution with 256 gray levels and size 576 × 768 are considered from abc imaging centre located in Vijayawada, Andhra Pradesh. Finally tested the proposed method on images taken from online available databases.
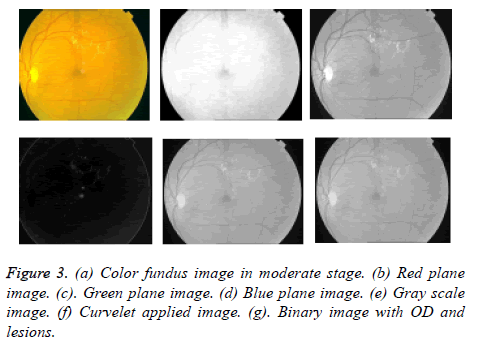
First we extracted green channel from color fundus image so that exudates appear brighter in this channel compared to other blue and red channels shown in Figures 3b-3d. Now on gray scale image shown in Figure 3e curvelet transform is applied. The curvelet transform decomposes image into several subbands with a given scale and orientation shown in Figure 3. The image is separated into a series of disjoint scales. Each subzone of each block analysed by ridgelet transform with rectangle size of 2p × 2-p/2 where k being a scale of curvelet transform. Basically, the assets of FFT are used by multiresolution curvelet transform in the spectral domain. While implementing FFT, the image and curvelet both are transformed into Fourier domain at a given scale and orientation. By applying inverse FFT we obtain a set of curvelet coefficients. Using Radon transform the curvelet coefficients can be obtained.
We use fast discrete curvelet transform via wrapping method for discrete translation of curvelet transform. In Fourier domain the curvelet coefficients Ck for each scale and angle is defined by
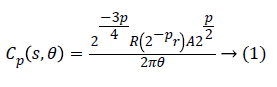
Where Cp is polar wedge supported by radial R and angular A windows.
In image segmentation, thresholding is the simplest technique [28]. The binary image is obtained by thresholding on gray scale image shown in Figures 4b and 4c. The stepwise FDCT via wrapping method is implemented in steps below.
A set of scales and directional bands coefficients Cp are obtained by applying FDCT via wrapping.
The threshold value is determined and compared with each directional band Cp.
Based on threshold value all coefficients are replaced.
Inverse transform is applied to convert image from transform domain to spectral domain.
The Peak signal to noise ratio is calculated for image quality measurement after curvelet transform implementation.
PSNR=20 × log10 (a/rms) → (2)
Where ‘a’ is the highest possible value of the signal or image (typically 255 or 1), and rms is the root mean square difference between two images. The PSNR measured in Decibel units (dB), which measure the ratio of the peak signal and the difference between two images.
Optic disk detection
Region Interest (ROI) is selected based on colormap range from the denoised image. Here color based region of interest is selected. The ROI is selected within intensity image and a binary image is obtained. ROI is pixel values that lie within the color map range Low (L) and High (H) as (L, H). Then binary image with zeroes outside the ROI and ones inside the ROI are obtained. The resultant binary image is having both OD and exudates shown in Figure 3g. The vasculature structure is eliminated automatically by selected thresholding values. Because compared to OD and bright lesion areas vessel parts having less intensity values.
BI=1 {if values selected inside ROI i.e., within (L, H)
BI=0 {otherwise → (3)
The Hough transform (HT) [29] is employed for the recognition of OD. HT is used to find the shapes of objects in image. It changes the image into parameter space. The parametric form for circle is given below,
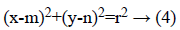
Where (m, n) is the centre of circle and r is radius that passes through (x, y).
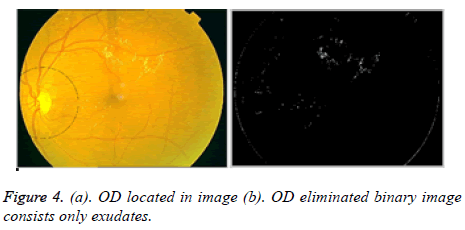
The Hough space is having three parameters; these are centre, radius, and collector. The collector is having a set of edge points consists circle of radius r. In Fundus image each pixel positions (x, y) of OD are detected. The circular shape is drawn around OD on color fundus image is located shown in Figure 4a.
Now the elimination of OD can be done based on selected thresholding values in the OD marked area. The OD lies in the left side or right side in the fundus image. Lesions occur near the damaged vessel structures mostly. So by selecting iteratively the intensity values from left and right side rows, columns based on image size 576 × 768. If the intensity value is less than or equal to 255 then put zero in the considered left and right side rows and columns in the image. Otherwise we put ones. Likewise OD part is eliminated.
OD eliminated ODe=0 {if intensity value I, 240 ≤ I ≤ 255
=1 {otherwise → (5)
In Figure 4b, only exudates presence is shown in the binary image.
Segmentation of exudates or bright lesions
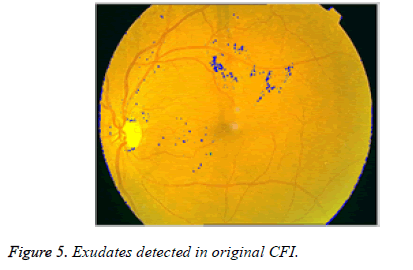
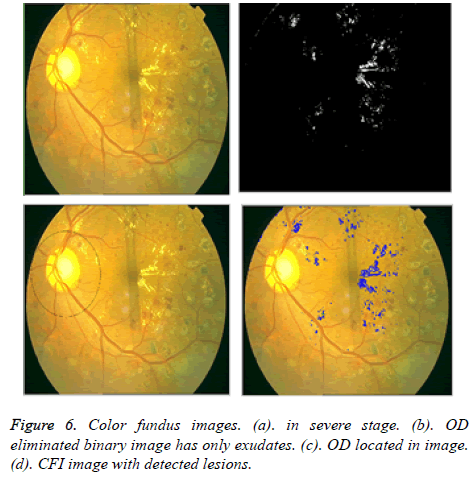
After the elimination of OD, binary image with only exudates area is shown in Figure 4b. Now based on Boundary tracing algorithm, we can identify the exudates in Color Fundus Image (CFI). Taking the binary image with only exudates as reference, boundaries can be drawn on damaged area particles in CFI. We trace the damaged area based on horizontal and vertical intensity values of original CFI as shown in Figure 5. The proposed system is tested for severe DR stage shown in Figure 6.
Feature extraction from detected lesions
The exudates detection phase extracts maximum possible number of lesions. By the adjustment of thresholding parameter, the missing of any exudates candidate is not possible in the segmentation stage. Exudates appear in yellow color or white color with varying sizes and shapes, but they are having sharp edges. To have the separation of exudates and non-exudates regions in retinal image, the feature set formation is necessary for an automated system. For classification, feature vector of lesion region is extracted. The feature vector for classification is shown below.
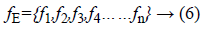
The detailed description for classification of lesions or nonlesions given below.
1. Area (f1): It is the sum of all pixels in detected exudate region.
2. Mean of intensity (f2): It gives the mean value of intensity in exudate region in green channel enhanced image.
3. Mean gradient magnitude (f3): To distinguish between blur and sharp edges, the pixels on edges are computed.
4. Entropy (f4): It gives the all pixel values in square block having exudate region and neighbouring pixels.
5. Energy (f5): Initially the summation of all pixel intensity values of lesion region done. Then these values are divided by total number of lesios pixel values.
6. Skewness (f6): The total pixels in square region having lesion candidate and the neighbouring pixel values.
Classification
In this phase SVM is implemented for DR severity grading. SVM is one of the supervised learning techniques [30-32]. The grading of DR is done based on the extracted features. SVM is having different hyper planes. To reach maximum margin, optimal hyper plane is selected for classification of different datasets. In this work, the SVM is applied twice. First, the separation of normal candidates from NPDR moderate and severe NPDR candidates is done. Next step is separating grade 2 and grade 3 candidates for lesion detection.
Discussion
The proposed system is also tested on online available databases diaretdb0 and diaretdb1. These databases are having American and Africans fundus images. Fundus eye image color changes from person to person. So tested proposed method on db0 and db1 databases also. Diaretdb1 dataset is having total 89 images with 500 field of view. These images are separated into two groups for training and testing.
The diaretdb0 database comprises 130 color fundus images with 500 field of view. In this database normal images are 20 and 110 images containing various lesions. These are signs of the diabetic retinopathy.
The statistical measurements used for evaluating the performance of proposed system defined in terms of True Positives (TP), False Positives (FP), True Negatives (TN), False Negatives (FN). The three parameters to measure accuracy are defined as:
Sensitivity (sen)=TP/(TP+FN)
Specificity (spe)=TN/ (TN+FP)
Accuracy (acc)=(sen+spe)/2
TP-Number of abnormal images correctly recognized as abnormal.
TN-Number of normal images correctly recognized as normal.
FP-Number of normal images incorrectly recognized as abnormal.
FN-Number of abnormal images incorrectly recognized as normal.
To know the diagnosis performance we have to measure the sen, spe, and acc parameters.
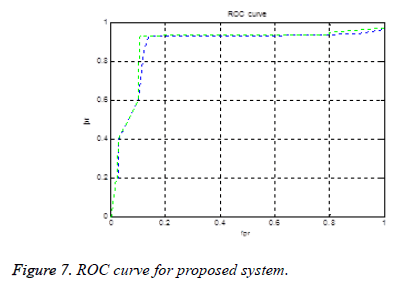
Figure 7 shows the ROC (receiver operating characteristic curve) curve. If an ROC curve shows AUC=1, then it is perfect diagnosis. If it shows 0.5, then it is worst case. In the proposed work obtained AUC=0.973. The measure of accuracy is proportional to the AUC.
Table 2 shows the performance with previous existing methods in terms of Sensitivity (in %), Specificity (in %), Accuracy (in %).
| Technique% | Sensitivity% | Specificity% | Accuracy |
|---|---|---|---|
| Alireza [8] | 96 | 94.6 | 95.3 |
| Wang [18] | 100 | 70 | 85 |
| Zhang [21] | 97 | 96 | 96.5 |
| Fleming [22] | 95 | 84.6 | 89.8 |
| Sinthanayothin [23] | 88.5 | 99.7 | 94.1 |
| Usher [24] | 95.1 | 46.3 | 70.7 |
| Walter [20] | 92.74 | 100 | 96.37 |
| Philip [25] | 90.5 | 67.4 | 78.95 |
| Abramoff [26] | 84 | 64 | 74 |
| Ahmed [36] | 94.9 | 100 | 97.45 |
| Osareh [19] | 93 | 94.1 | 93.55 |
| Haniza [35] | 94.25 | 99.2 | 96.75 |
| Sohini [33] | 100 | 83.16 | 91.58 |
| Deepak [34] | 100 | 90 | 95 |
| Proposed method | 100 | 94.2 | 97.53 |
Table 2. Comparison of performance of previous existing techniques (%).
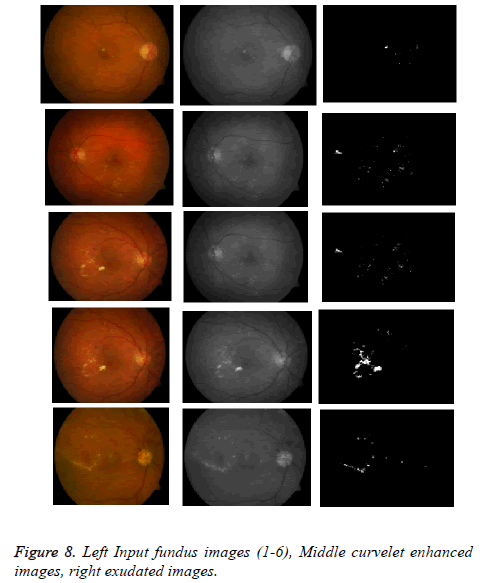
The proposed method tested on various images from online available datasets shown in Figure 8.
Conclusions
The proposed approach accomplished retinal image analysis for grading DR using SVM classifier. This system consists of pre-processing, OD detection, exudates region detection and DR grading based on classification. A detailed feature vector set depending on properties of exudates is obtained for each candidate exudates region for classification. The SVM classifier graded the input image into various categories. First proposed techniques tested on few images which were taken from the abc imaging centre located in Vijayawada, Andhra Pradesh. The recommended methodology is also tested on two online available retinal image databases, i.e. diaretdb1and diaretdb0 and succeeded for the recognition of exudates. Fundus eye image color changes from person to person and it is the main criteria, for the segmentation of exudates by choosing the threshold value. Our system attained a higher value of accuracy 97.53% compared to previous existing techniques. The efficiency of suggested method can be improved further by using a more accurate method for OD detection and by using the combination of classifiers. The results yield that the employed technique can be used in an automated medical system for grading of DR.
References
- International Diabetes Federation. Public health foundation of India. Data from the fact sheet diabetes in India. Fact sheet 2013.
- Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet 2010; 376: 124-136.
- Mohamed Q, Gillies MC, Wong TY. Management of diabetic retinopathy: a systematic review. J Am Med Assoc 2007; 298: 902-916.
- American Academy of Ophthalmology Retina Panel. Preferred practice pattern guidelines. Diab Retinop 2010.
- Ahmad MH, Nugroho H, Izhar LI, Nugroho HA. Analysis of retinal fundus images for grading of diabetic retinopathy severity. Med Biol Eng Comput 2011; 49: 693-700.
- Ram K, Joshi GD, Sivaswamy J. A successive clutter-rejection-based approach for early detection of diabetic retinopathy. IEEE Trans Biomed Eng 2011; 58: 664-673.
- Quellec G, Lamard M, Josselin PM, Cazuguel G, Cochener B. Optimal wavelet transform for the detection of microaneurysms in retina photographs. IEEE Trans Med Imaging 2008; 27: 1230-1241.
- Osareh A, Shadgar B, Markham R. A computational-intelligence-based approach for detection of exudates in diabetic retinopathy images. IEEE Trans Inf Technol Biomed 2009; 13: 535-545.
- Hatanaka Y, Nakagawa T, Hayashi Y, Kakogawa M, Sawada A, Kawase K, Hara T, Fujita H. Improvement of automatic haemorrhages detection methods using brightness correction on fundus images. SPIE Med Imag 2008; 6915: 5429.
- Akram UM, Khan SA. Automated detection of dark and bright lesions in retinal images for early detection of diabetic retinopathy. J Med Syst 2012; 36: 3151-3162.
- Walter T, Klein JC, Massin P, Erginay A. A contribution of image processing to the diagnosis of diabetic retinopathy-detection of exudates in color fundus images of the human retina. IEEE Trans Med Imag 2002; 21: 1236.
- Acharya UR, Chua CK, Ng EY, Yu W, Chee C. Application of higher order spectra for the identification of diabetes retinopathy stages. J Med Syst 2008; 32: 481-488.
- Lee SC, Lee ET, Wang Y, Klein R, Kingsley RM. Computer classification of non-proliferative diabetic retinopathy. Arch Ophthalmol 2005; 123: 759-764.
- Yun WL, Rajendra Acharya U, Venkatesh YV, Chee C, Min LC, Ng EYK. Identification of different stages of diabetic retinopathy using retinal optical images. Inform Sci 2008; 178: 106-121.
- Vallabha D, Dorairaj R, Namuduri K, Thompson H. Automated detection and classification of vascular abnormalities in diabetic retinopathy. IEEE Sig Sys Comp 2004; 2: 1625-1629.
- Tan NM, Wong DWK, Liu J, Ng WJ, Zhang Z, Lim JH, Tan Z, Tang Y, Li H, Lu S, Wong TY. Automatic detection of the macula in the retinal fundus image by detecting regions with low pixel intensity. Int Conf Biomed Pharmac Eng IEEE Singapore 2009; 9: 1-5.
- Acharya UR, Lim CM, Ng EY, Chee C, Tamura T. Computer-based detection of diabetes retinopathy stages using digital fundus images. Proc Inst Mech Eng H 2009; 223: 545-553.
- Wang H, Hsu W, Goh KG, Lee M. An effective approach to detect lesions in colour retinal images. Proc IEEE Conf Comp Vis Patt Recogn 2000; 181-187.
- Osareh A, Mirmehdi M, Thomas B, Markham R. Comparative exudate classification using support vector machines and neural networks. Int Conf Med Imag Comp Assi Interv 2002; 413-420.
- Walter T, Massin P, Erginay A, Ordonez R, Jeulin C. Automatic detection of microaneurysms in color fundus images. Med Image Anal 2007; 11: 555-566.
- Zhang X, Chutatape O. Detection and classification of bright lesions in colour fundus images. Int Conf Imag Proc 2004; 1: 139-142.
- Fleming AD, Philip S, Goatman KA, Williams GJ, Olson JA. Automated detection of exudates for diabetic retinopathy screening. Phys Med Biol 2007; 52: 7385-7396.
- Sinthanayothin C, Boyce JF, Williamson TH, Cook HL, Mensah E. Automated detection of diabetic retinopathy on digital fundus images. Diabet Med 2002; 19: 105-112.
- Usher D, Dumskyj M, Himaga M, Williamson TH, Nussey S. Automated detection of diabetic retinopathy in digital retinal images: a tool for diabetic retinopathy screening. Diabet Med 2004; 21: 84-90.
- Philip S, Fleming AD, Goatman KA, Fonseca S, McNamee P. The efficacy of automated disease/no disease grading for diabetic retinopathy in a systematic screening programme. Br J Ophthalmol 2007; 91: 1512-1517.
- Abràmoff MD, Niemeijer M, Suttorp-Schulten MS, Viergever MA, Russell SR. Evaluation of a system for automatic detection of diabetic retinopathy from color fundus photographs in a large population of patients with diabetes. Diabetes Care 2008; 31: 193-198.
- Candes LD, Donoho D, Ying L. Fast discrete curvelet transforms. Multiscale Model Simul 2006; 5: 861-899.
- Gonzalez RC, Woods RE. Thresholding in digital image processing. (Pearson Edn.) 2002.
- Akram MU, Khan A, Iqbal K, Butt WH. Retinal images: optic disk localization and detection. ICIAR 2010; 40-49.
- Vapnik V. Statistical learning theory. Springer 1998.
- Vapnik V, Golowich S, Smola A. Support vector method for function approximation, regression estimation, and signal processing. Adv Neur Inform Proc Sys 1997; 9: 281-287.
- Akram MU, Tariq A, Khan SA, Javed MY. Automated detection of exudates and macula for grading of diabetic macular edema. Comput Methods Programs Biomed 2014; 114: 141-152.
- Sohini K, Koozekanani DD . DREAM-Diabetic Retinopathy Analysis Using Machine Learning. IEEE J Biomed Health Inform 2014; 18.
- Deepak KS, Sivaswamy J. Automatic assessment of macular edema from color retinal images. IEEE Trans Med Imaging 2012; 31: 766-776.
- Yazid H, Arof H, Isa HM. Automated identification of exudates and optic disc based on inverse surface thresholding. J Med Syst 2012; 36: 1997-2004.
- Reza AW, Eswaran C, Dimyati K. Diagnosis of diabetic retinopathy: automatic extraction of optic disc and exudates from retinal images using marker controlled watershed transformation. J Med Sys 2011; 35: 1491-1501.