Research Article - Biomedical Research (2018) Volume 29, Issue 18
Complex networks of interaction of genes located in the critical region of Down syndrome expressed in the normal human brain
Dianora Fajardo1*, Karla Vinasco2, Julio C Montoya3, Jose M Satizabal3, Adalberto Sanchez3 and Felipe García-Vallejo4
1Department of Biochemistry, Institución Universitaria Escuela Nacional del Deporte, Cali, Colombia
2LABIOMOL, Universidad del Valle, Cali, Colombia
3Department of Physiological Sciences, School of Basic Sciences, Universidad del Valle, Colombia
4Scientific Director of the Laboratory of Molecular Biology and Pathogenesis, Department of Physiological Sciences, School of Basic Sciences, Universidad del Valle, Cali, Colombia
- *Corresponding Author:
- Dianora Fajardo
Department of Biochemistry
Institución Universitaria Escuela Nacional del Deporte, Colombia
Accepted on July 12, 2018
DOI: 10.4066/biomedicalresearch.29-18-690
Visit for more related articles at Biomedical ResearchAbstract
The quantification and analysis of the human transcriptome allows expanding the knowledge of the genomic functioning, especially in body’s parts as complex and important as the human brain. In this way, in silico studies offer the possibility to extract and analyse information contained in databases, at the level of gene expression along the different brain structures. This study aimed to correlate the transcription levels of 38 genes located in the critical region of the chromosome 21 associated with Down syndrome with the cerebral localization and its intervention in the correct operation of different brain substructures. To carry out this, the expression profiles of these genes along 24 substructures of the brain cores and 18 of the Limbic lobe were done, from gene expression data of microarray experiments of DNA, available in the database of the Atlas of the brain of the "Allen Institute for Brain Sciences". It was determined a differential expression of these genes along the analysed structures, in addition to register higher levels of overall transcription in certain areas of the brain, which appear to be associated with different processes of learning and memory. The differential transcription was correlated with the cerebral localization and its potential functional role.
Keywords
Basal ganglia, Limbic lobe, DNA microarrays, Critical region of Down syndrome, PCP4, KCNJ6, DYRK1A.
Introduction
The human brain has a wide variety of cells, each of those ones has a single morphophysiology, functionality and connectivity [1,2]. These properties are largely the result of unique combinations of expressed gene products, and its precise regulation is what keeps to a large cerebral homeostasis degree. This approach is useful for understanding the functional circuitry of the nervous system, and thus generates new knowledge about the relationship between genes, brain and behavior [3-5]. Therefore, the analysis of gene expression profiles provides huge information about brain connectivity and its relationship with the higher cognitive functions. As an example there is the influence of genetic factors in understanding the normal brain development and mental disorders [5-8]. Consequently, the cellular diversity of the brain requires a focus on understanding the functional genomics of the nervous system, leading to the incorporation of all these modalities in an overall analysis having the potential to improve the discovery and highlighting its importance in comparison with methods that analyse each mode separately. Data associations between these three approaches are relevant in the study of diseases, since gene expression in the brain plays a key role in the flow of information between brain networks and performing cognitive tasks [3,5].
For that reason, the inherent importance of brain homeostasis and the complexity involved in the development and maintenance of the nervous system can result in a series of neuropathology that lead to changes in brain structure such as decreasing different types of motricity, cognitive impairment, among others [6]. Down Syndrome (DS), which is caused by the total or partial presence of three copies of chromosome 21, is becoming the most frequent aneuploidy leading to vary degrees of cognitive impairment [9-11]. On the other hand, studies of segmental trisomies have allowed to characterize an area within the chromosome called “Critical Region of the Down Syndrome” (DSCR), which is located at the distal end of the long arm of chromosome 21 (21q22.1-22.3), and which contains possible candidate genes whose imbalance of dose could induce marked cognitive deficit, like the other pathologies and traits associated [12-17].
However, the involvement of the DSCR as the sole cause of the symptoms of DS is still a matter of debate. Several studies suggest that this region plays a major role in the genetic interactions that would be related to the pathogenesis of DS [18-20]. In spite of this, the expression of the genes which are found within this region is not fully known in the brain. Therefore, their study in normal human brains could provide a better understanding of their participation in the regulation of all processes that must be performed for proper operation. Additionally, a comprehensive approach would make significant correlations between gene expression and regulation, the function of the nervous system and the resulting phenotype. This would be too informative for neurogenetic and study of brain diseases, especially in the neurological disorders associated with the DS.
Actually, few studies focus on the functional analysis correlation of gene expression in the brain. Moreover, the techniques that have been used, usually cover large regions of the brain creating a difficulty in interpreting data in the substructures, or generally made by a gene at a time, leaving patterns of expression of many genes uncharacterized. As a result, Atlas Allen Human Brain Project has adopted a global approach for understanding the structural and genetic architecture of the brain by generating gene expression profiles obtained from DNA microarrays from post-mortem human brains. In this context, it is possible to extract complete and detailed information on those levels of transcription in different brain structures that can be found in the database of free access of the Allen Brain Atlas [21]. This database contains anatomical and genomic information from human brains, which is supplemented with extra information and set of visualization tools and data mining. Microarray experiments include data of more than 62,000 probes, covering 93% of the 21,245 genes consigned, from which it is possible to obtain information on thousands transcriptional gene referenced with Entrez gene codes.
This study was designed to construct an in silico model of the expression profiles of 38 genes located in the critical region of DS. Data were based to correlate them with the functionality of a healthy human brain of a 55 y old male donor, besides, a network of expression and interaction of these genes with others that expression was built. The results allowed us to approach to a systemic model of expression that can be modified bioinformatically to extrapolate it to what happens in other diseases of the brain, providing a powerful tool for its understanding.
Materials and Methods
Data collection
The gene expression levels were calculated from the z-score values of 38 genes DSCR (Annex 1) in different substructures of the basal ganglia and the limbic lobe. These were obtained from the graphical display of the database of the human brain the Allen Institute for Brain Sciences. All procedures used for collecting data are reported extensively in the technical report “Allen Institute for Brain Sciences” [23].
| Structure name | Symbol |
|---|---|
| Brain nuclei | |
| Caudate nuclei head-left | CNH-Left |
| Caudate nuclei head-right | CNH-Right |
| Caudate nuclei tail-left | CNT-Left |
| Caudate nuclei tail-right | CNT-Right |
| Putamen left | Pu-Left |
| Putamen right | Pu-Right |
| Nucleus accumulus left | NA-Left |
| Nucleus accumulus right | NA-Right |
| Globus pallidum external left | GPE-Left |
| Globus pallidum external right | GPE-Right |
| Globus pallidum internal left | GPI-Left |
| Globus pallidum internal right | GPI-Right |
| Septal nuclei left | SptN-Left |
| Septal nuclei right | SptN-Right |
| Basal forebrain substantia innominata left | BFbSI-Left |
| Basal forebrain substantia innominata right | BFbSI-Right |
| Claustrum left | C-Left |
| Claustrum right | C-Right |
| Amigdala transition zone | ATZ |
| Amigdala basolateral left | ABL-Left |
| Amigdala basomedia right | ABM-Right |
| Central amigdala nucleus | ACN |
| Corto medial amigdala | ACM |
| Left nucleus amigdala | ALN |
| Limbic lobe | |
| Short insular gyrus | SIG |
| Cyngulate girus, frontal part | CgGf |
| Cyngulate girus, parietal part | CgG-Parietal |
| Dental gyrus left | DG-Left |
| Central area 1 left | CA1-Left |
| Central area 1 right | CA-1 Right |
| Central area 2 left | CA2-Left |
| Central area 2 right | CA-2 Right |
| Central area 3 left | CA3-Left |
| Central area 3 right | CA-3 Right |
| Central area 4 left | CA-4-Left |
| Central area 4 right | CA-4 Right |
| Subiculum left | S-Left |
| Subiculum right | S-Right |
| Parahipocampus gyrus left | PHG-Left |
| Parahipocampus gyrus -cos-left | PHG-cos-Left |
| Parahipocampus gyrus right | PHG-Right |
| Parahipocampus gyrus -cos-right | PHG-cos-Right |
Annex 1. Names and symbols of different substructures in limbic lobe and cerebral nuclei. The names were extracted from the Allen Institute for Brain Sciences.
In all cases, the data of each gene of available experiments using different probes was obtained. Standard values (z score) of the expression levels through 24 substructures of the brain nuclei and 18 from the Limbic Lobe (Annex 2) were recorded. These values were recorded in electronic sheets in Excel format for further analysis. Three values were taken at three different points of each substructure and the average z score was regarded as final data.
| Gene | Common | Entrez ID | SIG | CgGf | CgG-Par | DG-Left |
|---|---|---|---|---|---|---|
| RCAN1 | NM_004414.5 | 1827 | 0.139 | -0.3595 | -0.228 | -1.03 |
| PSMG1 | NM_203433.1 | 8624 | 1.131 | 0.2285 | -2.309 | 1.292 |
| DSCR3 | NM_006052.1 | 10311 | 1.018 | -0.2485 | -0.233 | 0.711 |
| DSCR4 | NM_005867.2 | 10281 | 1.436 | -0.2535 | 1.096 | -0.25 |
| DSCR6 | NM_018962.1 | 53820 | 0.572 | -0.249 | 0.43 | 0.764 |
| DSCR8 | NR_026838.1 | 84677 | 0.387 | -0.291 | 1.114 | 0.844 |
| PIGP | NM_153681.1 | 51227 | 0.373 | -0.149 | -2.107 | 2.122 |
| CLIC6 | NM_053277.1 | 54102 | 0.09 | -0.316 | 0.563 | -1.035 |
| BACE2 | NM_012105.3 | 25825 | -2.501 | 0.0305 | -0.676 | -2.431 |
| BRWD1 | NM_018963.3 | 54014 | 0.08 | 0.0605 | 0.095 | 1.178 |
| DSCAM | NM_001389.3 | 1826 | 0.836 | 0.011 | 0.189 | 0.79 |
| DYRK1A | NM_130438.1 | 1859 | -0.101 | 0.5235 | 0.503 | 0.663 |
| ERG | NM_001136154.1 | 2078 | -0.204 | -0.341 | 0.072 | -0.643 |
| ETS2 | NM_005239.4 | 2114 | -0.427 | -0.501 | 0.145 | 0.889 |
| KCNJ6 | NM_002240.2 | 3763 | 1.76 | 0.257 | 0.086 | 1.574 |
| RUNX1 | NM_001122607.1 | 861 | 0.387 | -0.571 | -0.101 | -1.355 |
| SH3BGR | NM_001001713.1 | 6450 | 1.151 | 0.7375 | -0.112 | 0.185 |
| SIM2 | NM_005069.2 | 6493 | 0.306 | -0.365 | -0.105 | -1.081 |
| CLDN14 | NM_012130.2 | 23562 | -0.012 | 0.107 | -0.283 | 0.126 |
| TTC3 | NM_001001894.1 | 7267 | 0.437 | 0.551 | 0.465 | 1.175 |
| SON | NM_138927.1 | 6651 | 0.162 | -0.184 | -0.023 | 0.489 |
| HLCS | NM_000411.4 | 3141 | 0.179 | -0.257 | 0.300 | 1.808 |
| KCNJ15 | NM_002243.3 | 3772 | -0.668 | 0.453 | -0.081 | 0.010 |
| HMGN1 | NM_004965.6 | 3150 | -0.345 | -0.094 | -0.328 | 0.530 |
| WRB | NM_004627.4 | 7485 | 0.401 | 0.699 | -0.782 | -0.269 |
| LCA5L | NM_152505.2 | 150082 | 0.224 | 0.088 | 1.671 | -0.057 |
| C21orf88 | NR_026542.1 | 114041 | -0.480 | -0.039 | -0.959 | 0.507 |
| B3GALT5 | NM_033173.1 | 10317 | -0.776 | 0.138 | -0.924 | 0.257 |
| TMEM1 | NM_003274.3 | 7109 | -0.627 | -0.313 | -0.645 | 0.433 |
| IGSF5 | NM_001080444.1 | 150084 | -0.828 | 0.078 | -1.009 | -0.323 |
| PCP4 | NM_006198.2 | 5121 | -0.237 | -0.532 | -0.822 | -0.042 |
| CSTB | NM_000100.2 | 1476 | 0.202 | 0.618 | -0.271 | -1.553 |
| TMEM50B | NM_006134.4 | 757 | -0.144 | -0.230 | -0.464 | -0.858 |
| PTTG1IP | NM_004339.2 | 754 | 0.741 | 0.070 | -0.814 | -0.563 |
| TIAM1 | NM_003253.1 | 7074 | -0.849 | -0.899 | -0.171 | 2.290 |
| PRMT2 | NM_206962.1 | 3275 | -0.013 | -0.110 | -0.430 | -0.171 |
| Gene | Common | Entrez ID | CA3-left | CA-3 Right | CA4-left | CA-4 Right |
| RCAN1 | NM_004414.5 | 1827 | 0.056 | 0.106 | 2.641 | -0.137 |
| PSMG1 | NM_203433.1 | 8624 | 1.166 | 1.28 | 2.823 | -1.401 |
| DSCR3 | NM_006052.1 | 10311 | 0.204 | 0.484 | 0.808 | -0.976 |
| DSCR4 | NM_005867.2 | 10281 | 0.373 | -0.008 | 2.99 | -0.104 |
| DSCR6 | NM_018962.1 | 53820 | 0.788 | 0.151 | 2.237 | 1.182 |
| DSCR8 | NR_026838.1 | 84677 | 1.743 | 0.886 | 2.721 | 0.713 |
| PIGP | NM_153681.1 | 51227 | 1.587 | 0.349 | 1.877 | 2.006 |
| CLIC6 | NM_053277.1 | 54102 | -0.285 | -0.627 | 0.37 | -0.175 |
| BACE2 | NM_012105.3 | 25825 | -1.167 | -1.363 | 0.975 | -0.495 |
| BRWD1 | NM_018963.3 | 54014 | 0.357 | 0.213 | -1.598 | -0.154 |
| DSCAM | NM_001389.3 | 1826 | 0.494 | -0.249 | 1.917 | 1.463 |
| DYRK1A | NM_130438.1 | 1859 | -0.093 | 0.687 | -1.506 | -1.329 |
| ERG | NM_001136154.1 | 2078 | 0.197 | -0.49 | 1.694 | 0.293 |
| ETS2 | NM_005239.4 | 2114 | -0.689 | -0.923 | -0.125 | 0.247 |
| KCNJ6 | NM_002240.2 | 3763 | 1.05 | 1.2 | -1.101 | 1.01 |
| RUNX1 | NM_001122607.1 | 861 | -0.713 | -0.173 | 1.753 | 0.657 |
| SH3BGR | NM_001001713.1 | 6450 | 0.35 | 0.561 | 0.273 | 0.44 |
| SIM2 | NM_005069.2 | 6493 | -0.09 | 0.011 | 1.959 | 0.76 |
| CLDN14 | NM_012130.2 | 23562 | 0.983 | -0.230 | -0.532 | 0.265 |
| TTC3 | NM_001001894.1 | 7267 | 1.744 | 1.642 | 1.470 | 1.470 |
| SON | NM_138927.1 | 6651 | -0.581 | -0.381 | -0.230 | -0.475 |
| HLCS | NM_000411.4 | 3141 | -0.086 | -0.457 | -0.123 | -0.451 |
| KCNJ15 | NM_002243.3 | 3772 | 1.133 | -0.131 | -0.558 | 0.282 |
| HMGN1 | NM_004965.6 | 3150 | -0.556 | -0.821 | -0.288 | -0.455 |
| WRB | NM_004627.4 | 7485 | -1.105 | 0.111 | -0.242 | -0.748 |
| LCA5L | NM_152505.2 | 150082 | 0.270 | -0.424 | -0.362 | -0.354 |
| C21orf88 | NR_026542.1 | 114041 | 1.453 | 1.421 | 1.167 | 1.509 |
| B3GALT5 | NM_033173.1 | 10317 | 1.402 | 0.127 | 0.081 | 0.265 |
| TMEM1 | NM_003274.3 | 7109 | 0.709 | -0.091 | 0.030 | 0.258 |
| IGSF5 | NM_001080444.1 | 150084 | 0.089 | 0.623 | 1.201 | -0.136 |
| PCP4 | NM_006198.2 | 5121 | -2.043 | -2.133 | -1.909 | -1.466 |
| CSTB | NM_000100.2 | 1476 | -0.686 | -1.037 | -0.192 | -1.277 |
| TMEM50B | NM_006134.4 | 757 | -1.411 | -0.288 | -0.602 | -0.898 |
| PTTG1IP | NM_004339.2 | 754 | -0.924 | -0.868 | -0.354 | -0.701 |
| TIAM1 | NM_003253.1 | 7074 | -0.279 | -0.631 | -0.479 | -0.096 |
| PRMT2 | NM_206962.1 | 3275 | -0.825 | -0.706 | -0.407 | -0.743 |
| Gene | Common | Entrez ID | PHG-R | PHG-cos-R | CNH-Left | CNH-Right |
| RCAN1 | NM_004414.5 | 1827 | 0.261 | 0.931 | 1.112 | 1.433 |
| PSMG1 | NM_203433.1 | 8624 | -0.305 | 0.457 | -0.589 | -1.794 |
| DSCR3 | NM_006052.1 | 10311 | -0.363 | -0.393 | 0.707 | -0.423 |
| DSCR4 | NM_005867.2 | 10281 | -0.354 | -0.031 | -0.433 | 0.216 |
| DSCR6 | NM_018962.1 | 53820 | -0.238 | -0.233 | 1.281 | 2.312 |
| DSCR8 | NR_026838.1 | 84677 | 0.095 | 1.576 | -0.749 | 0.793 |
| PIGP | NM_153681.1 | 51227 | -1.419 | 0.108 | 0.306 | -0.548 |
| CLIC6 | NM_053277.1 | 54102 | 2.147 | 1.22 | 0.849 | -0.397 |
| BACE2 | NM_012105.3 | 25825 | -1 | -0.327 | 0.169 | -0.164 |
| BRWD1 | NM_018963.3 | 54014 | 0.244 | 0.179 | -0.403 | -0.196 |
| DSCAM | NM_001389.3 | 1826 | 0.475 | 0.693 | -0.643 | -0.1475 |
| DYRK1A | NM_130438.1 | 1859 | 0.304 | 0.268 | -0.94 | -1.861 |
| ERG | NM_001136154.1 | 2078 | 0.594 | -0.424 | -0.025 | 0.317 |
| ETS2 | NM_005239.4 | 2114 | -0.417 | -0.085 | -0.952 | -1.439 |
| KCNJ6 | NM_002240.2 | 3763 | 0.489 | 0.165 | -2.365 | -2.283 |
| RUNX1 | NM_001122607.1 | 861 | -0.179 | -0.555 | 1.408 | 1.79 |
| SH3BGR | NM_001001713.1 | 6450 | 0.59 | 0.364 | 0.75 | 0.965 |
| SIM2 | NM_005069.2 | 6493 | -0.164 | -0.608 | 0.02 | 0.645 |
| CLDN14 | NM_012130.2 | 23562 | 0.797 | 0.035 | 0.285 | 0.633 |
| TTC3 | NM_001001894.1 | 7267 | 1.949 | 0.823 | -0.938 | -0.508 |
| SON | NM_138927.1 | 6651 | 0.339 | -0.493 | 0.478 | 0.242 |
| HLCS | NM_000411.4 | 3141 | 0.793 | -0.003 | 2.027 | 2.049 |
| KCNJ15 | NM_002243.3 | 3772 | 0.849 | -0.073 | 0.268 | 0.553 |
| HMGN1 | NM_004965.6 | 3150 | 0.781 | -0.015 | 0.664 | 1.308 |
| WRB | NM_004627.4 | 7485 | 0.776 | -0.265 | -0.032 | -0.433 |
| LCA5L | NM_152505.2 | 150082 | 0.473 | 0.518 | 1.770 | 0.294 |
| C21orf88 | NR_026542.1 | 114041 | 0.040 | 0.782 | 2.238 | 3.418 |
| B3GALT5 | NM_033173.1 | 10317 | 0.180 | 0.505 | 2.611 | 3.416 |
| TMEM1 | NM_003274.3 | 7109 | -0.206 | -0.284 | 0.654 | 0.979 |
| IGSF5 | NM_001080444.1 | 150084 | -1.103 | -0.869 | 2.376 | 2.394 |
| PCP4 | NM_006198.2 | 5121 | -1.141 | -0.147 | 3.066 | 3.867 |
| CSTB | NM_000100.2 | 1476 | -0.054 | 0.944 | 1.688 | 1.104 |
| TMEM50B | NM_006134.4 | 757 | -0.192 | -0.570 | 0.167 | 0.230 |
| PTTG1IP | NM_004339.2 | 754 | 0.617 | 0.684 | 1.041 | 1.404 |
| TIAM1 | NM_003253.1 | 7074 | -0.251 | -0.431 | 0.295 | 0.396 |
| PRMT2 | NM_206962.1 | 3275 | 0.030 | 0.157 | -1.436 | -1.944 |
| Gene | Common | Entrez ID | NA-Left | NA-Right | GPE-Left | GPE-Right |
| RCAN1 | NM_004414.5 | 1827 | 0.518 | 0.353 | 2.669 | 2.12 |
| PSMG1 | NM_203433.1 | 8624 | -0.198 | -0.237 | -1.912 | -1.563 |
| DSCR3 | NM_006052.1 | 10311 | -1.337 | -0.935 | 1.089 | 0.614 |
| DSCR4 | NM_005867.2 | 10281 | 1.824 | 0.04 | -0.064 | -0.09 |
| DSCR6 | NM_018962.1 | 53820 | -1.268 | -1.834 | 0.102 | 0.174 |
| DSCR8 | NR_026838.1 | 84677 | 0.724 | -0.348 | 0.543 | 0.748 |
| PIGP | NM_153681.1 | 51227 | 1.971 | 0.633 | 1.055 | 1.131 |
| CLIC6 | NM_053277.1 | 54102 | -0.692 | 0.335 | 1.797 | 0.752 |
| BACE2 | NM_012105.3 | 25825 | -0.457 | 0.258 | 2.444 | 2.176 |
| BRWD1 | NM_018963.3 | 54014 | 0.282 | 0.369 | -0.644 | -0.429 |
| DSCAM | NM_001389.3 | 1826 | 0.493 | 0.662 | 0.107 | 0.637 |
| DYRK1A | NM_130438.1 | 1859 | -0.328 | -0.616 | -1.358 | -1.694 |
| ERG | NM_001136154.1 | 2078 | -0.372 | 0.465 | 2.089 | 1.617 |
| ETS2 | NM_005239.4 | 2114 | 0.665 | 0.517 | -1.152 | -1.27 |
| KCNJ6 | NM_002240.2 | 3763 | -0.618 | -0.177 | -1.778 | -1.569 |
| RUNX1 | NM_001122607.1 | 861 | 0.286 | 0.49 | 0.687 | 1.482 |
| SH3BGR | NM_001001713.1 | 6450 | 0.833 | 0.688 | 0.77 | 1.241 |
| SIM2 | NM_005069.2 | 6493 | 0.285 | -0.461 | 0.983 | 1.313 |
| CLDN14 | NM_012130.2 | 23562 | 0.019 | -0.507 | 0.663 | 0.971 |
| TTC3 | NM_001001894.1 | 7267 | 0.028 | -0.539 | -1.828 | -1.589 |
| SON | NM_138927.1 | 6651 | 0.119 | 0.864 | 0.879 | 1.192 |
| HLCS | NM_000411.4 | 3141 | 1.349 | 0.169 | 1.705 | 1.717 |
| KCNJ15 | NM_002243.3 | 3772 | 0.119 | 0.118 | 0.683 | 0.197 |
| HMGN1 | NM_004965.6 | 3150 | 0.849 | 0.944 | 0.793 | 0.885 |
| WRB | NM_004627.4 | 7485 | 0.297 | 0.532 | 0.955 | 1.493 |
| LCA5L | NM_152505.2 | 150082 | 0.359 | 0.152 | 0.673 | 0.551 |
| C21orf88 | NR_026542.1 | 114041 | 1.415 | 1.150 | 0.893 | 0.536 |
| B3GALT5 | NM_033173.1 | 10317 | 0.574 | 0.258 | -0.012 | -0.198 |
| TMEM1 | NM_003274.3 | 7109 | -0.146 | -0.854 | 1.312 | 1.226 |
| IGSF5 | NM_001080444.1 | 150084 | 1.273 | 0.944 | 1.142 | 1.000 |
| PCP4 | NM_006198.2 | 5121 | 1.689 | 1.727 | 0.917 | 0.822 |
| CSTB | NM_000100.2 | 1476 | 0.445 | 0.752 | 1.407 | 1.522 |
| TMEM50B | NM_006134.4 | 757 | 0.235 | 0.661 | 0.932 | 1.167 |
| PTTG1IP | NM_004339.2 | 754 | 0.547 | 0.144 | 2.952 | 2.643 |
| TIAM1 | NM_003253.1 | 7074 | 0.576 | 0.324 | 0.104 | -0.381 |
| PRMT2 | NM_206962.1 | 3275 | -0.156 | 0.382 | 0.693 | 0.768 |
| Gene | Common | Entrez ID | BFbSI-L | BFbSI-R | C-Left | C-Right |
| RCAN1 | NM_004414.5 | 1827 | 0.339 | 0.888 | -1.268 | -0.295 |
| PSMG1 | NM_203433.1 | 8624 | -0.357 | -1.262 | -0.008 | -0.075 |
| DSCR3 | NM_006052.1 | 10311 | 0.069 | 0.247 | -1.554 | -0.976 |
| DSCR4 | NM_005867.2 | 10281 | -0.48 | -0.274 | 0.785 | 0.542 |
| DSCR6 | NM_018962.1 | 53820 | 0.318 | -0.471 | 1.397 | 0.668 |
| DSCR8 | NR_026838.1 | 84677 | -0.366 | -0.358 | 1.896 | 0.677 |
| PIGP | NM_153681.1 | 51227 | -0.397 | -0.122 | -3 | -0.34 |
| CLIC6 | NM_053277.1 | 54102 | 0.001 | -0.472 | 0.549 | -0.4 |
| BACE2 | NM_012105.3 | 25825 | 0.962 | -0.024 | -0.803 | 0.42 |
| BRWD1 | NM_018963.3 | 54014 | -0.492 | -0.269 | 0.381 | 0.861 |
| DSCAM | NM_001389.3 | 1826 | 0.908 | 0.67 | 1.005 | 0.999 |
| DYRK1A | NM_130438.1 | 1859 | -0.648 | -0.661 | -0.782 | 0.348 |
| ERG | NM_001136154.1 | 2078 | -1.04 | 0.744 | 0.812 | 0.873 |
| ETS2 | NM_005239.4 | 2114 | -1.182 | -0.191 | 0.859 | 0.392 |
| KCNJ6 | NM_002240.2 | 3763 | -1.357 | -1.297 | -0.526 | -0.427 |
| RUNX1 | NM_001122607.1 | 861 | -0.387 | -0.245 | 1.299 | 0.532 |
| SH3BGR | NM_001001713.1 | 6450 | 0.014 | 0.715 | 0.224 | 0.013 |
| SIM2 | NM_005069.2 | 6493 | -0.112 | 0.797 | 0.863 | 1.395 |
| CLDN14 | NM_012130.2 | 23562 | -0.457 | -0.435 | 2.331 | 1.152 |
| TTC3 | NM_001001894.1 | 7267 | -0.387 | -0.479 | 0.033 | -0.076 |
| SON | NM_138927.1 | 6651 | -0.131 | -0.471 | -0.180 | -0.024 |
| HLCS | NM_000411.4 | 3141 | -0.659 | 0.685 | -1.000 | -0.859 |
| KCNJ15 | NM_002243.3 | 3772 | -0.831 | 0.222 | -0.155 | 0.149 |
| HMGN1 | NM_004965.6 | 3150 | 0.619 | -0.296 | 0.035 | -0.482 |
| WRB | NM_004627.4 | 7485 | 0.621 | 0.362 | 0.020 | 0.004 |
| LCA5L | NM_152505.2 | 150082 | 0.434 | -0.533 | -0.320 | 0.330 |
| C21orf88 | NR_026542.1 | 114041 | 0.387 | 0.172 | -1.228 | -1.098 |
| B3GALT5 | NM_033173.1 | 10317 | 0.024 | 0.313 | -0.031 | 0.068 |
| TMEM1 | NM_003274.3 | 7109 | -0.372 | -0.344 | 0.889 | 1.019 |
| IGSF5 | NM_001080444.1 | 150084 | 0.942 | 0.528 | -1.210 | -1.394 |
| PCP4 | NM_006198.2 | 5121 | 1.241 | 1.255 | -0.416 | -0.324 |
| CSTB | NM_000100.2 | 1476 | 0.953 | 1.087 | 0.600 | 0.044 |
| TMEM50B | NM_006134.4 | 757 | 0.547 | 0.274 | -1.117 | -1.051 |
| PTTG1IP | NM_004339.2 | 754 | 0.253 | 0.097 | 0.048 | -0.169 |
| TIAM1 | NM_003253.1 | 7074 | 0.158 | -0.206 | -1.369 | -1.539 |
| PRMT2 | NM_206962.1 | 3275 | 0.785 | 0.718 | 0.715 | 0.827 |
Annex 2. Standard values (z score) of the expression levels through 24 substructures of the brain nuclei and 18 from the limbic lobe.
Protein interaction network and cluster analysis
A network of interaction of proteins encoded by 36 of the DSCR genes with other human proteins in different databases was built through a Cytoscape program version 3.1.1 [24]. Besides, a hierarchical clustering was performed based on the z score of these 36 genes in each cerebral substructure. The test conditions used were the plugin clusterMaker Algorithm maximum link by pairs and metric distance metric Pearson correlation.
Statistical analysis
For ranking each expression of genes in different brain substructures, a Principal Component Analysis (PCA) for both the limbic lobe and the brain nuclei was performed by reducing the R space of 38 collinear variables to 6 main components (R38>R6) in each area. This analysis was performed using IBM SPSS 20.0.0 program [25].
Results
DSCR overall transcription of genes in the brain
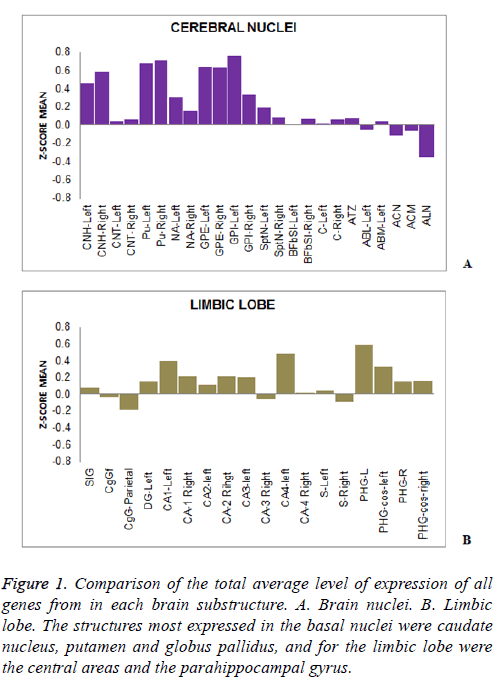
A differential expression of 38 genes DSCR along different brain substructures, specific and dependent on the associated substructure brain, was determined. In turn it was confirmed the existence of regulation of gene expression dependent on the physiology of each brain area (Figure 1). From the brain areas studied, the caudate nucleus, putamen and globus pallidus are substructures that had a higher level of expression by the majority of genes in the basal nuclei (Figure 1A). The most expressed areas in the limbic lobe were the central and the parahippocampal gyrus (Figure 1B).
Figure 1: Comparison of the total average level of expression of all genes from in each brain substructure. A. Brain nuclei. B. Limbic lobe. The structures most expressed in the basal nuclei were caudate nucleus, putamen and globus pallidus, and for the limbic lobe were the central areas and the parahippocampal gyrus.
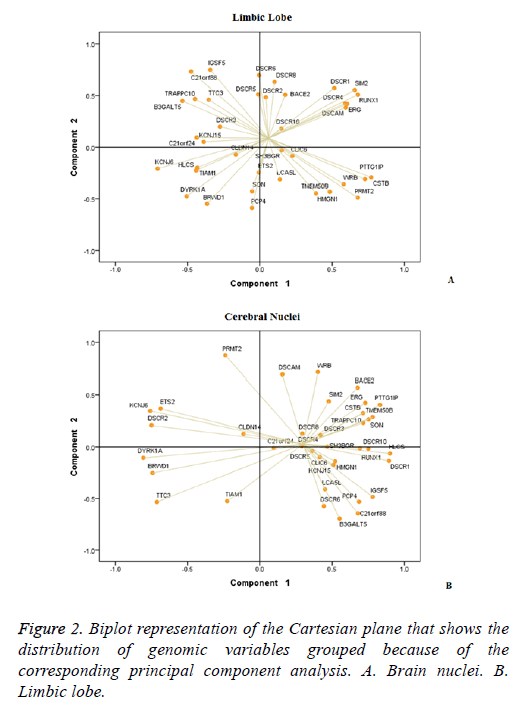
The PCA allowed reducing to six components the 38 genes DSCR in each structure. In brain, nuclei represented 79.89% of the variance while the limbic lobe 76.03%. It was observed that the distribution of the 38 DSCR genes was differential in the two brain structures (Figure 2). In brain nuclei component 1 was the most complex, which includes 19 genes, in contrast to the limbic lobe where 19 of the 38 genes DSCR distributed between components 1 and 2 (Table 1).
| Brain structure | Component | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| Cerebral nuclei | KCNJ15 | DSCR1 | DSCAM | DSCR4 | DSCR2 | CLIC6 | DSCR3 |
| LCA5L | DSCR5 | ETS2 | DSCR6 | BRWD1 | DYRK1A | TIAM1 | |
| C21orf88 | DSCR10 | WRB | DSCR8 | C21orf24 | |||
| B3GALT5 | BACE2 | PRMT2 | SIM2 | HMGN1 | |||
| TRAPPC10 | ERG | CLDN14 | KCNJ6 | ||||
| IGF5 | RUNX1 | TTC3 | |||||
| PCP4 | SH3BGR | ||||||
| CSTB | SON | ||||||
| TMEM50B | HLCS | ||||||
| PTTG1IP | |||||||
| Limbic lobe | DSCAM | DSCR1 | DSCR10 | BRWD1 | KCNJ6 | DSCR3 | |
| DSCR4 | DSCR2 | CLDN14 | ETS2 | SH3BGR | CLIC6 | ||
| RUNX1 | DSCR6 | KCNJ15 | SON | DYRK1A | |||
| ERG | DSCR8 | C21orf24 | HLCS | PCP4 | |||
| WRB | DSCR5 | LCA5L | HMGN1 | ||||
| SIM2 | BACE2 | B3GALT5 | TRAPPC10 | ||||
| CSTB | TTC3 | TIAM1 | |||||
| TMEM50B | C21orf88 | ||||||
| PTTG1IP | IGSF5 | ||||||
| PRMT2 | |||||||
Table 1. Discrimination on the weight of each of the variables within six components.
In addition, comparative analysis of graphics Biplot establishes two groups of genes having a statistically significant correlation for each structure. At Limbic lobe, the first association is composed by the IGSF5, C21orf88, TRAPPC10, TTC3, B3GALT5, DSCR3, KCNJ15 and C21orf24; and the second by the DSCR1, SIM2, RUNX1, ERG, DSCAM and DSCR4 (Figure 2A) genes. While in brain nuclei, the stronger association included the BACE2, SIM2, ERG, CSTB, PTTG1IP, TMEM50B, TRAPPC10 genes (Figure 2B).
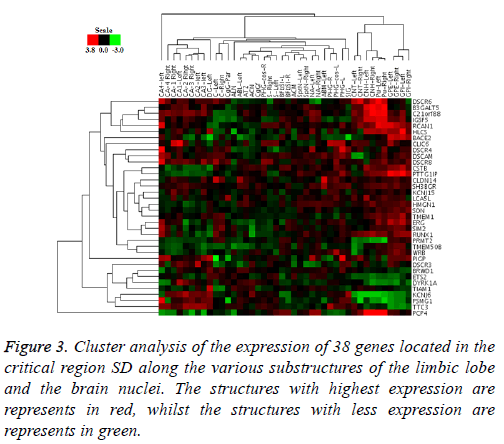
The PCA analysis showed that gene co-expression varies depending on the brain area. Both in brain nuclei and the limbic lobe, two groups presented where the genes that compose them are highly correlated, but all the associations that occur differ in both structures. Similarly, the cluster analysis showed the same pattern of co-expression (Figure 3), leaving also areas of high and low expression mentioned before, and the existence of an opposite expression of certain genes in these two areas.
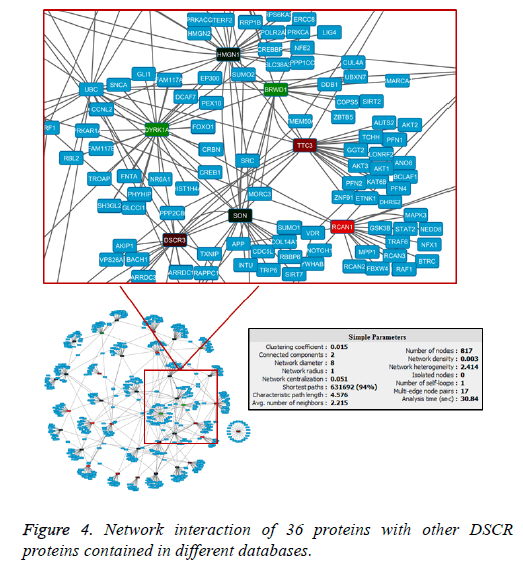
Protein interaction network
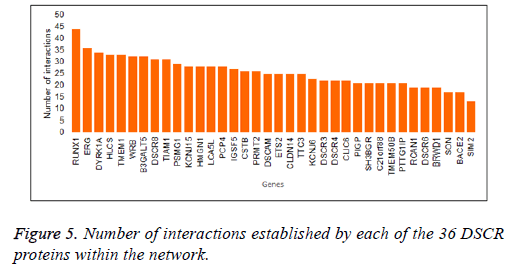
The interaction network showed a master node that stands out within seven DSCR proteins interacting within them and with a variable number of others human proteins. The network showed that in the primary node DYRK1A gene product is found (Figure 4). It had one of the highest values of interactions within the network (Figure 5), and interacts directly with another six proteins DSCR: HMGN1, BRWD1, TTC3, RCAN1, ARE and DCSR3.
In the same way, the top ten GO categories with highest statistical significance in terms of biological process were analysed. As shown in Table 2, most of the proteins studied in the interaction network regulate positively and/or negatively various biological and metabolic processes, mechanisms of apoptosis, modification of macromolecules, among others.
| GO-ID | Description | P-value | Cluster frequency | Total frequency |
|---|---|---|---|---|
| 48523 | Negative regulation of cellular process | 5.35E-17 | 179/702 (25.4%) | 1840/14274 (12.8%) |
| 48519 | Negative regulation of biological process | 1.32E-15 | 186/702 (26.4%) | 2016/14274 (14.1%) |
| 6464 | Protein modification process | 1.49E-13 | 148/702 (21.0%) | 1527/14274 (10.6%) |
| 43412 | Macromolecule modification | 3.63E-13 | 152/702 (21.6%) | 1608/14274 (11.2%) |
| 10941 | Regulation of cell death | 1.13E-12 | 99/702 (14.1%) | 868/14274 (6.0%) |