Research Article - Journal of Genetics and Molecular Biology (2022) Volume 6, Issue 2
Comparative structures and evolution of sterol-C5-desaturase (SC5D) genes and proteins: A major role in cholesterol biosynthesis.
Roger S Holmes*
Department of Medicine, Griffith Research Institute for Drug Design (GRIDD) and School of Environment and Science, Griffith University, Nathan, QLD, Australia
- *Corresponding Author:
- Roger S Holmes
Department of Medicine
Griffith Research Institute for Drug Design
Griffith University, Nathan, QLD
4111 Australia
Phone: +61-410-583-348
Email: r.holmes@griffith.edu.au
Received: 07-Feb-2022, Manuscript No. AAGMB-22-53649; Editor assigned: 09-Feb-2022, PreQC No. 23-Feb-2022 (PQ); Reviewed: 14-Mar-2022, QC No. AAGMB-22-53649; Published: 28-Mar-2022, DOI:10.35841/aagmb-6.2.106
Citation: Holmes RS. Comparative structures and evolution of sterol-C5-desaturase (SC5D) genes and proteins: A major role in cholesterol biosynthesis. J Genet Mol Bio 2022;6(2):106
Abstract
Sterol-C5-desaturase (SC5D) (EC 1.14.21.6) catalyses a reaction in the cholesterol biosynthetic pathway, converting lathosterol into 7-dehydrocholesterol. The enzyme is broadly distributed in human tissues, with highest levels in liver and brain. Bioinformatic methods were used to predict the amino acid sequences, transmembrane and gene locations for SC5D genes and encoded proteins using data from several vertebrate and invertebrate genome projects. Multiple histidine rich motifs (HR[G/F]LHH, HKPHH and HHTDHH) were conserved and four transmembrane regions were usually observed for the vertebrate SC5D protein sequences examined. Vertebrate SC5D genes predominantly contained 4 coding exons transcribed on either the positive or negative DNA strands. Evidence is presented for chromosomally duplicated SC5D genes for the Xenopus laevis genome. Vertebrate SC5D protein subunits shared 66-100% sequence identities and exhibited sequence alignments and identities for amino acid residues as well as extensive conservation of predicted transmembrane structures. Phylogenetic analyses demonstrated the relationships and evolutionary origins of the vertebrate SC5D genes which were related to an invertebrate SC5D gene observed in nematode and fruit fly genomes.
Keywords
Vertebrates, Amino acid sequence, SC5D, Sterol-C5-desaturase, Evolution, Phylogeny, Cholesterol biosynthesis.
Introduction
Sterol-C5-desaturase (SC5D) (EC 1.14.21.6) (also designated as lathosterol oxidase, lathosterol dehydrogenase and lathosterol desaturase) (SC5DL; S5DES; ERG3) is a membrane-bound enzyme in liver and other tissues of the body which catalyzes the penultimate reaction in the cholesterol biosynthetic pathway, converting lathosterol into 7-dehydrocholesterol by introducing a C5-6 double bond into lathosterol [1-3]. Mutations of the human SC5D gene may result in the Smith-Lemli-Optiz syndrome, which is a frequently occurring autosomal recessive developmental disease resulting in mental retardation, facial dysmorphisms, and other congenital anomalies, as well as the accumulation of cholesterol intermediates [4-7].
Structures for SC5D genes and cDNA sequences have been determined, including human (Homo sapiens) [3,8,9] and mouse (Mus musculus) [10,11]. Human SC5D spans 20.96 kbps, comprises 4 exons and is localized on chromosome 11 on the direct strand [12]. SC5D is broadly expressed in human and mouse tissues, with highest levels in liver and brain, where the enzyme contributes predominantly to cholesterol biosynthesis. Studies of Sc5d¯/Sc5d¯ knock out mice have shown that these mice were still born with elevated lathosterol and decreased cholesterol levels and exhibited craniofacial defects, including cleft palates as well as limb patterning defects [10].
This paper reports gene structures and amino acid sequences for several vertebrate SC5D genes and proteins, the predicted secondary structures for vertebrate SC5D protein subunits, and the structural and evolutionary relationships for these genes and enzymes.
Materials and Methods
Vertebrate SC5D gene and protein identification
Protein BLAST analyses used web tools and SC5D amino acid sequences previously reported (Table 1) [8,9,13]. Protein sequence databases for several vertebrate genomes were examined, including human (Homo sapiens); mouse (Mus musculus); opossum (Monodelphis domestica); platypus (Ornithorhynchus anatinus); chicken (Gallus gallus); lizard (Anolis carolinensis); tropical toad (Xenopus tropicalis); African clawed toad (Xenopus laevis); zebrafish (Danio rerio); coelacanth (Latimeria chalumnae); nematode (Pristionchus pacificus); and fruit fly (Drosophila melanogaster) [14-24].
BLAST analyseswere subsequently undertaken for each the predicted vertebrateSC5D amino acid sequences using the UC Santa Cruz web [25]. To obtain the predicted locations for each of the vertebrate SC5D genes, including predicted exon boundary locations and gene sizes. Structures for human and mouse SC5D isoforms (splicing variants) were obtained using the AceView website to examine predicted gene and protein structures [26].
Predicted structures and properties of vertebrate SC5D protein subunits
Predictions of transmembrane helices for vertebrate SC5D subunits were obtained using the TMHMM web server v. 2.0
Comparative human (sc5d) tissue expression
The UCSC Genome [25]. was used to examine comparative human tissue SC5D expression.
Phylogenetic studies and sequence divergence
Alignments and phylogeny analyses of vertebrate SC5D were undertaken using the phylogeny.fr web browser (Table 1) [27].
| Gene | Animal | Species | Gene | Transcript ID | Exons | Gene | Amino | Identity % |
|---|---|---|---|---|---|---|---|---|
| Location | *prediction | (Strand) | size | acids | human | |||
| SC5D | human | Homo sapiens | 11:121303376-121307509 | NM_006918 | 4 (+) | 4134 | 299 | 100 |
| Sc5d | mouse | Mus musculus | 9:42255345-42259891 | BAA33730.1 | 4 (-) | 4547 | 299 | 84 |
| SC5D | cow | Bos taurus | 15:31882315-31886419 | *XP_005216026.2 | 4 (+) | 4105 | 299 | 85 |
| SC5D | opossum | Monodelphis domestica | 4:227383420-227389596 | *XP_001380248.2 | 4 (-) | 6177 | 299 | 74 |
| SC5D | platypus | Ornithorhynchus anatinus | ^DS18116v1:240342-248431 | *XP_028930992.1 | 4 (-) | 8090 | 299 | 75 |
| SC5D | chicken | Gallus gallus | 24:3560554-3563750 | *XP_015153662.1 | 4 (-) | 3197 | 288 | 66 |
| SC5D | lizard | Anolis carolensis | Lga:2222395-2231447 | *XP_003222854.1 | 4 (+) | 9053 | 293 | 66 |
| SC5D | tropical frog | Xenopus tropicalis | 7:88269461-88277246 | CX849787 | 4 (+) | 7786 | 290 | 68 |
| SC5DS | clawed frog | Xenopus laevis | 7S:54461781-54466717 | *XP_018083286.1 | 4 (+) | 4937 | 290 | 69 |
| SC5DL | clawed frog | Xenopus laevis | 7L:71071125-71075126 | NP_001087137 | 4 (-) | 4002 | 287 | 70 |
| SC5D | zebra fish | Danio rerio | 21:24531949-24534481 | NP_001004630.1 | 4 (-) | 2533 | 300 | 66 |
| SC5D | shark | Callorhinchus milii | ^KI636257:127617-131679 | *XP_007909349.1 | 4 (-) | 4063 | 301 | 66 |
| SC5D | coelacanth | Latimeria chalumnae | ^JH126563:455076-464346 | *XP_005986182.1 | 4 (+) | 9271 | 299 | 67 |
| SC5D | sea hare | Aplysia californica | ^Sc1016:160717-172702 | *XP_005110921.1 | 5 (-ve) | 11986 | 303 | 62 |
| SC5D | purple sea urchin | Strongylocentrus purpuratus | ^Sc65907:33352-38703 | *XP_030831120.1 | 3 (-ve) | 5352 | 305 | 60 |
| SC5D | nematode | Pristionchus pacificus | ^Chr:112653770-11265678 | KAF8384310.1 | 11 (-ve) | 2409 | 302 | 22 |
| SC5D | fruit fly | Drosophila melanogaster | X:13676915-13677818 | CG11162 | 2 (+) | 904 | 278 | 27 |
Table 1: Vertebrate and invertebrate SC5D genes and proteins.
Results and Discussion
Alignments of SC5D subunit sequences
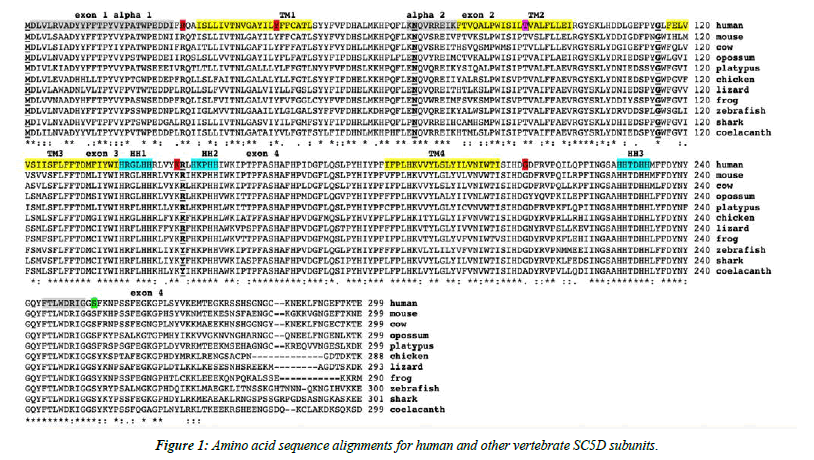
The amino acid sequences for human (Homo sapiens) and mouse (Mus musculus) SC5D subunits and for other vertebrate SC5D subunits are shown in Figure 1. These alignments showed between 66-100% sequence identities, suggesting that these are products of the same family of genes and proteins (Table 1). In contrast, invertebrate SC5D sequences exhibited much lower sequence identities with the human sequence, with nematode (Pristionchus pacificus) and fruit fly (Drosophila melanogaster) SC5D sequences showing identities of 22% and 27% respectively with the human SC5D sequence (Table 1). The amino acid sequences for human and other mammalian SC5D enzymes contained 299 residues whereas other vertebrate and invertebrate SC5D subunits contained between 278 residues (fruit fly) and 305 (purple sea urchin) amino acids (Figure 1, Table 1). These differences were predominantly explained by changes in the number of C-terminal amino acids (Figure 1).
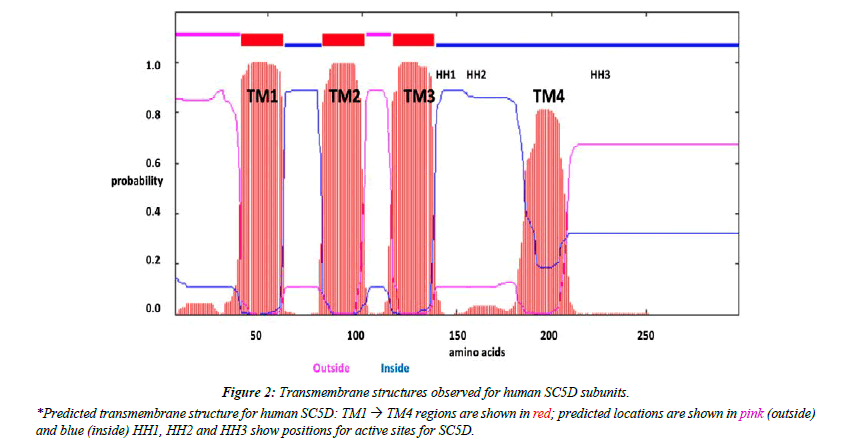
Predicted transmembrane and active site sequences of vertebrate SC5D subunits
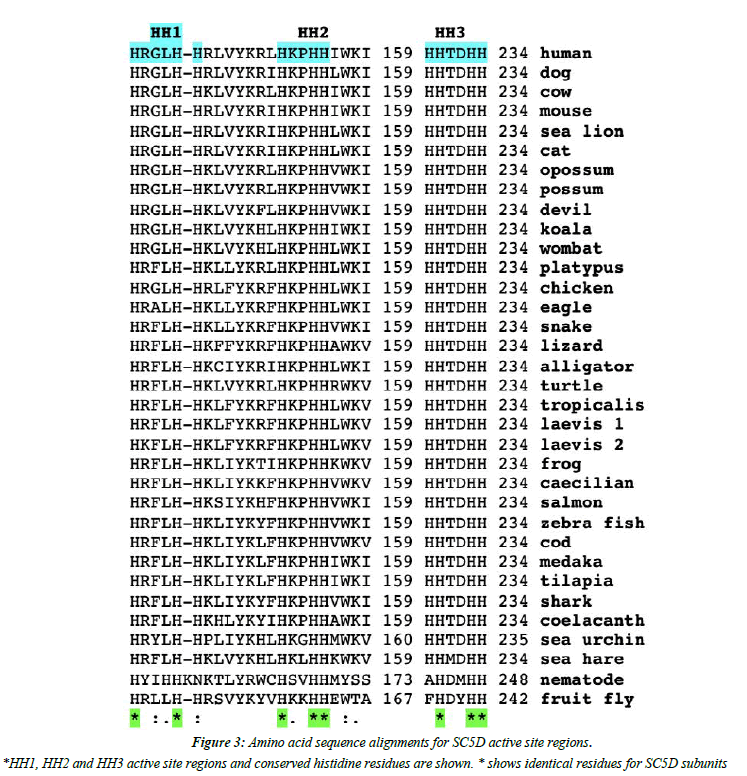
Analyses of predicted secondary and transmembrane structures for vertebrate SC5D sequences were examined which were consistent with four transmembrane regions (designated as TM1-4) and two alpha helical regions (alpha 1 and 2) (Figure 1 and Figure 2). Similar structures were observed for each of the vertebrate SC5D subunits examined. For human SC5D, the following secondary structures were observed: alpha helix 1 (residues 2-27); TM1 (36-58); alpha helix 2 (70-78); TM2 (79-101); TM3 (116-138); and TM4 (182-204), with intracellular locations predicted for inside (residues 1-34; 100-114; 205-299) and outside (58-76; 138- 181) the endoplasmic reticulum membrane (Figure 2). The active site for vertebrate SC5D is associated with conserved histidine regions (designated as HH1, HH2 and HH3) which were centrally located near TM3 and TM4. These residues are likely to play a role in providing ligands for a proposed catalytic Fe center for enzymes responsible for desaturating and hydroxylating reactions, such as SC5D; FA2H (fatty acid α-hydroxylase); and fatty acid desaturases (FADS1; FADS2; FADS3) [28-30]. Figure 3 shows the comparative putative histidine active site sequences for all vertebrate and invertebrate SC5D sequences examined demonstrating conservation of the following histidine residues (human SC5D sequence used): His138; His142; His151; His154; His155; His229; His232; His233, supporting an essential role for these amino acids in vertebrate and invertebrate SC5D catalysis. This proposal is further substantiated by site directed mutagenesis studies of Arabidoopsis thaliana Δ7-sterol-C5(6)- desaturase (5-DES) for which mutations of the corresponding active site histidine residues have generated 2-20% activity levels of the wild-type enzyme [28].
Gene locations and exon structures for SC5D genes and proteins
Table 1 summarizes the predicted locations for vertebrate and invertebrate SC5D genes based upon BLAT interrogations of the corresponding genomes using the reported sequences for human, mouse and rat SC5D [2,7-9] and predicted sequences for other vertebrate and invertebrate SC5D genes and proteins. Human and mouse SC5D genes were located on chromosomes 11 and 9, respectively, and contained 4 coding exons in each case (Table 1). The clawed toad (Xenopus laevis) genome showed evidence of duplicated SC5D genes, with predicted SC5DS and SC5DL genes being localized on separate chromosomes (chr7S and chr7L, respectively), apparently reflecting an allotetraploid origin from two diploid progenitor Xenopus species around 18 million years ago [20]. In contrast, the related diploid tropical toad (Xenopus tropicalis) genome provided evidence of a single SC5D gene on chr7 (Table 1). Invertebrate SC5D genes examined also contained multiple coding exons: sea hare (Aplysia californica), 5 coding exons; purple sea urchin (Strongylcentrus purpuratus), 3 coding exons; nematode (Pristionchus pacificus), 11 coding exons; and the fruit fly genome containing a SC5D gene, with 2 coding exons localized on the X chromosome (Table 1).
Figure 1 summarizes the predicted exon start sites for human, mouse and several other vertebrate SC5D genes with each having 4 exons, in identical positions to those reported for the human SC5D and mouse Sc5d genes [3,8,9]. The four transmembrane regions (TM1-TM4) were distributed within each of the encoded exons with the three putative histidine active site sequences positioned between exons 3 and 4 (HH1/ HH2) and after TM4 (HH3). Lathosterolosis, an inborn error of the cholesterol biosynthetic metabolic pathway, has been reported clinically in human and mouse genomes due to SC5D deficiency [7,10,31-33]. In human clinical studies of the impact of SC5D mutations, this condition has been described as the Smith-Lemli-Opitz syndrome (SLOS) [5]. resulting in the formation of abnormal sterols and chondrodysplasia punctata [6]. SLOS disease variants (also referred to as LATHOS variants) have been reported involving amino acid substitutions (29R→Q; 46Y→S; and 211 G→D), resulting in SC5D deficiencies [10,31]. Studies with Sc5d (-/-) mice have shown that the neonates were stillborn with elevated lathosterol levels, decreased cholesterol levels and craniofacial defects; Consistent with phenotypes reported for human SC5D defective gene patients [10,33]. and supporting an essential role for the SC5D gene in normal mammalian development (Figure 1).
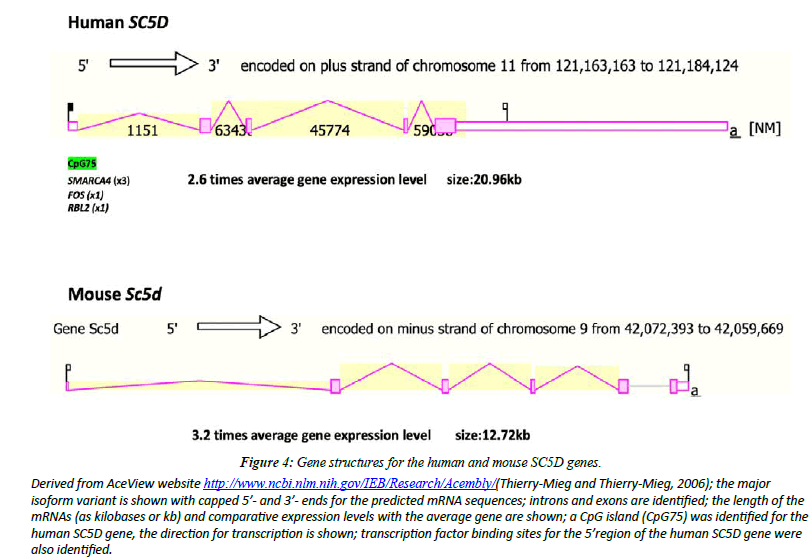
Figure 4 illustrates the structures of mRNAs for human SC5D and mouse Sc5d transcripts [26]. The human SC5D genome sequence contained several predicted transcription factorbinding sites (TFBS) and a CpG island (CpG75) SMARCA4 has been recognized as a potential transcription factor binding site for the human SC5D gene reulating gene expression by altering chromatin structure, particularly in proliferating mammary epithelial cells [34]. FOS [35] and RBL2 [36] have also been identified as likely sites of regulating expression of the human SC5D gene. The occurrence of the CpG island within the SC5D gene may reflect a role in regulating gene expression [37]. Contributing to a higher gene expression level reported for human SC5D (x2.6 times higher) and mouse Sc5d (3.2 times higher).
Figure 4: Gene structures for the human and mouse SC5D genes.
Derived from AceView website http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/(Thierry-Mieg and Thierry-Mieg, 2006); the major isoform variant is shown with capped 5’- and 3’- ends for the predicted mRNA sequences; introns and exons are identified; the length of the mRNAs (as kilobases or kb) and comparative expression levels with the average gene are shown; a CpG island (CpG75) was identified for the human SC5D gene, the direction for transcription is shown; transcription factor binding sites for the 5’region of the human SC5D gene were also identified.
Comparative human sc5d tissue expression
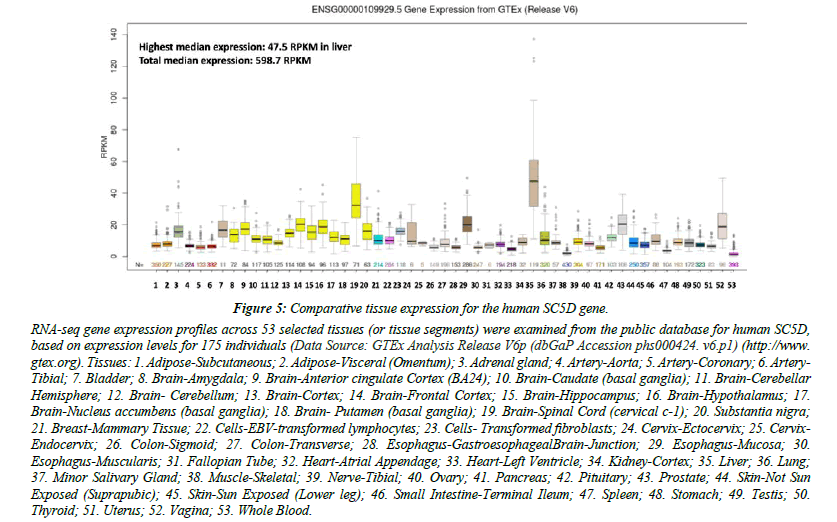
Figure 5 shows comparative gene expression for various human tissues obtained from RNA-seq gene expression profiles for the human SC5D gene obtained for 53 selected tissues or tissue segments for 175 individuals [35] (Data Source: GTEx Analysis Release V6p (dbGaP Accession phs000424.v6.p1). These data supported a much higher level of tissue expression for human SC5D in liver and the spinal cord, with highest median expression observed in liver [38- 41]. The presence of multiple TFBS within the SC5D gene promoter (SMARCA4, FOS and RBL2) may contribute to this high level in expression level.
Figure 5: Comparative tissue expression for the human SC5D gene.
RNA-seq gene expression profiles across 53 selected tissues (or tissue segments) were examined from the public database for human SC5D, based on expression levels for 175 individuals (Data Source: GTEx Analysis Release V6p (dbGaP Accession phs000424. v6.p1) (http://www.gtex.org). Tissues: 1. Adipose-Subcutaneous; 2. Adipose-Visceral (Omentum); 3. Adrenal gland; 4. Artery-Aorta; 5. Artery-Coronary; 6. Artery- Tibial; 7. Bladder; 8. Brain-Amygdala; 9. Brain-Anterior cingulate Cortex (BA24); 10. Brain-Caudate (basal ganglia); 11. Brain-Cerebellar Hemisphere; 12. Brain- Cerebellum; 13. Brain-Cortex; 14. Brain-Frontal Cortex; 15. Brain-Hippocampus; 16. Brain-Hypothalamus; 17. Brain-Nucleus accumbens (basal ganglia); 18. Brain- Putamen (basal ganglia); 19. Brain-Spinal Cord (cervical c-1); 20. Substantia nigra; 21. Breast-Mammary Tissue; 22. Cells-EBV-transformed lymphocytes; 23. Cells- Transformed fibroblasts; 24. Cervix-Ectocervix; 25. Cervix-Endocervix; 26. Colon-Sigmoid; 27. Colon-Transverse; 28. Esophagus-GastroesophagealBrain-Junction; 29. Esophagus-Mucosa; 30. Esophagus-Muscularis; 31. Fallopian Tube; 32. Heart-Atrial Appendage; 33. Heart-Left Ventricle; 34. Kidney-Cortex; 35. Liver; 36. Lung; 37. Minor Salivary Gland; 38. Muscle-Skeletal; 39. Nerve-Tibial; 40. Ovary; 41. Pancreas; 42. Pituitary; 43. Prostate; 44. Skin-Not Sun Exposed (Suprapubic); 45. Skin-Sun Exposed (Lower leg); 46. Small Intestine-Terminal Ileum; 47. Spleen; 48. Stomach; 49. Testis; 50. Thyroid; 51. Uterus; 52. Vagina; 53. Whole Blood.
Phylogeny and divergence of vertebrate and invertebrate SC5D sequences
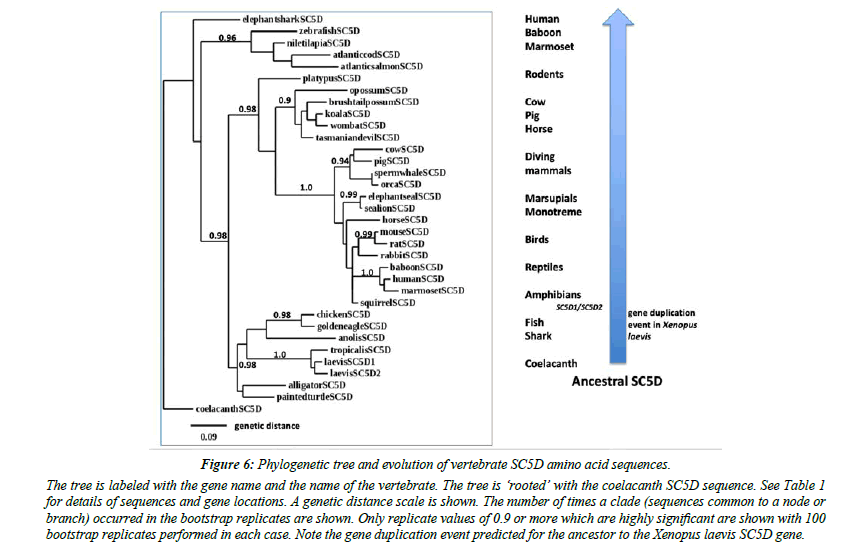
A phylogenetic tree (Figure 6) was calculated by the progressive alignment of human and other vertebrate SC5D amino acid sequences. The phylogram was ‘rooted’ with a coelacanth SC5D sequence and showed clustering of the SC5D sequences into several distinct vertebrate groups, including primates (human, baboon and marmoset); other eutherian mammals (ruminants, diving mammals, seals, rodents); marsupial and monotreme species); birds and reptiles; amphibians; fish; and a shark and a coelacanth species. In addition, Xenopus laevis SC5D1 and SC5D2 sequences showed clustering with the Xenopus tropicalis sequence examined, which is consistent with these genes being products of a recent duplication event during Xenopus evolution [20].
Figure 6: Phylogenetic tree and evolution of vertebrate SC5D amino acid sequences..
The tree is labeled with the gene name and the name of the vertebrate. The tree is ‘rooted’ with the coelacanth SC5D sequence. See Table 1 for details of sequences and gene locations. A genetic distance scale is shown. The number of times a clade (sequences common to a node or branch) occurred in the bootstrap replicates are shown. Only replicate values of 0.9 or more which are highly significant are shown with 100 bootstrap replicates performed in each case. Note the gene duplication event predicted for the ancestor to the Xenopus laevis SC5D gene.
Conclusion
In conclusion, the results of the present study indicate that vertebrate SC5D genes and encoded SC5D enzymes represent a distinct sterol desaturase gene and enzyme family which share key conserved sequences and structures with transmembrane structures and associated histidine boxes consistent with these serving as active sites and/or involved with metal ion binding. SC5D expression is highly expressed in liver and the spinal cord which may be facilitated by transcription factor binding sites, SMARCA4, FOS and RBL2, which are localized in the 5’ region of the SC5D gene on the human genome his study also reports that the Xenopus laevis genome contains at least two ‘SC5D-like’ genes (designated as SC5D1 and SC5D2), which were located in tandem on separate chromosomes (chr7S and chr7L) of the Xenopus laevis genome. Phylogeny studies using several vertebrate SC5D subunits indicated that SC5D genes have apparently appeared early in vertebrate evolution prior to the teleost fish common ancestor more than 500 million years ago.
Acknowledgements
The advice and support of Dr Laura Cox, Wake Forest Medical Center, Winston Salem NC USA is gratefully acknowledged.
References
- Koroly MJ, Dempsey ME. Synthesis of delta 5,22-cholestadien-3 beta-ol from delta 5,7,22-cholestatrien-3 beta-ol by a liver enzyme. Lipids. 1981;16(10):755-8.
- Kawata S, Trzaskos JM, Gaylor JL. Microsomal enzymes of cholesterol biosynthesis from lanosterol. Purification and characterization of delta 7-sterol 5-desaturase of rat liver microsomes. J Biol Chem. 1985; ;260(11):6609-17.
- Nishi S, Nishino H, Ishibashi T. CDNA cloning of the mammalian sterol C5-desaturase and the expression in yeast mutant. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression. 2000;1490(1-2):106-8.
- Waterham HR, Wijburg FA, Hennekam RC, et al. Smith-Lemli-Opitz syndrome is caused by mutations in the 7-dehydrocholesterol reductase gene. J Am Hum Genet. 1998;63(2):329-38.
- Witsch-Baumgartner M, Fitzky BU, Ogorelkova M, et al. Mutational spectrum in the Δ7-sterol reductase gene and genotype-phenotype correlation in 84 patients with Smith-Lemli-Opitz syndrome. J AmHumGenet. 2000;66(2):402-12.
- Ruan B, Tsai J, Wilson WK, et al. Aberrant pathways in the late stages of cholesterol biosynthesis in the rat: origin and metabolic fate of unsaturated sterols relevant to the Smith-Lemli-Opitz syndrome. JLipid Res. 2000;41(11):1772-82.
- Gaoua W, Wolf C, Chevy F, et al. Cholesterol deficit but not accumulation of aberrant sterols is the major cause of the teratogenic activity in the Smith-Lemli-Opitz syndrome animal model. JLipid Res. 2000;41(4):637-46.
- Moebius FF, Fitzky BU, Lee JN, et al. Molecular cloning and expression of the human Δ7-sterol reductase. ProcNatlAcadSci. 1998;95(4):1899-902.
- Sugawara T, Fujimoto Y, Ishibashi T. Molecular cloning and structural analysis of human sterol C5 desaturase. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2001;1533(3):277-84.
- Krakowiak PA, Wassif CA, Kratz L, et al. Lathosterolosis: An inborn error of human and murine cholesterol synthesis due to lathosterol 5-desaturase deficiency. HumMolGenet. 2003;12(13):1631-41.
- Makarova AM, Pasta S, Watson G, et al. Attenuation of UVR-induced vitamin D3 synthesis in a mouse model deleted for keratinocyte lathosterol 5-desaturase. J Steroid BiochemMolBiol. 2017;171:187-94.
- Lee BT, Barber GP, Benet-Pagès A, et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022;50(D1):D1115-22.
- Altschul F, Vyas V, Cornfield A, et al. Basic local alignment search tool. J Mol Biol. 1990; 215: 403-10.
- International Human Genome Sequencing Consortium. Correction: initial sequencing and analysis of the human genome. Nature. 2001;412(6846):565.
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695-716.
- Mikkelsen TS, Wakefield MJ, Aken B, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447(7141):167-77.
- Warren WC, Hillier LW, Graves JA, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453(7192):175.
- Alföldi J, Di Palma F, Grabherr M, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477(7366):587-91.
- Hellsten U, Harland RM, Gilchrist MJ, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328(5978):633-6.
- Session AM, Uno Y, Kwon T, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538(7625):336-43.
- Sprague J, Bayraktaroglu L, Clements D, et al. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581-5.
- Amemiya CT, Alföldi J, Lee AP, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496(7445):311-6.
- Dieterich C, Clifton SW, Schuster LN, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. NatGenet. 2008;40(10):1193-8.
- Adams MD, Celniker SE, Holt RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185-95.
- Karolchik D, Baertsch R, Diekhans M, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31(1):51-4.
- Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Bio. 2006;7(1):1-4.
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465-9.
- Taton M, Husselstein T, Benveniste P, et al. Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase studied by site-directed mutagenesis. Biochemistry. 2000;39:701-11.
- Zhu L, Koszelak-Rosenblum M, Connolly SM, et al. The crystal structure of an integral membrane fatty acid alpha-hydroxylase. J Biol Chem. 2015;290(50):29820-33.
- Cui J, Chen H, Tang X, et al. Delta6 fatty acid desaturases in polyunsaturated fatty acid biosynthesis: insights into the evolution, function with substrate specificities and biotechnological use. Appl Microbio Biotechnol. 2020;104:9947-63.
- Brunetti-Pierri N, Corso G, Rossi M, et al. Lathosterolosis, a novel multiple-malformation/mental retardation syndrome due to deficiency of 3β-hydroxysteroid-Δ5-desaturase. J AmHumGenet. 2002;71(4):952-8.
- Gondré-Lewis MC, Petrache HI, Wassif CA, et al. Abnormal sterols in cholesterol-deficiency diseases cause secretory granule malformation and decreased membrane curvature. J Cell Sci. 2006;119:1876-85.
- Jiang XS, Backlund PS, Wassif CA, et al. Quantitative proteomics analysis of inborn errors of cholesterol synthesis: identification of altered metabolic pathways in DHCR7 and SC5D deficiency. MolCell Proteomics. 2010;9(7):1461-75.
- Barutcu AR, Lajoie BR, Fritz AJ, et al. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016;26(9):1188-201.
- Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. J Europ Cancer. 2005;41(16):2449-61.
- Ullah F, Khan T, Ali N, et al. Promoter methylation status modulate the expression of tumor suppressor (RbL2/p130) gene in breast cancer. PLoS One. 2015;10(8):0134687.
- Saxonov V, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci. 2006;103(5):1412-7.
- Fitzky BU, Witsch-Baumgartner M, Erdel M, et al. Mutations in the Δ7-sterol reductase gene in patients with the Smith–Lemli–Opitz syndrome. Proc Natl Acad Sci. 1998;95(14):8181-6.
- GTEx Consortium, Ardlie KG, Deluca DS, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Sci. 2015;348(6235):648-60.
- Waterston RH, Pachter L. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520-62.
- Rigbolt KT, Prokhorova TA, Akimov V, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4(164):rs3.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref