Keywords
|
| Glutamate decarboxylase; Lactobacillus plantarum; Bioinformatics Analysis γ-aminobutyric acid. |
Introduction
|
| γ-Aminobutyric acid (GABA) is a non-protein amino acid that is widely distributed in nature and plays an important role as the major inhibitory neurotransmitter in the mammalian central nervous system [1]. It has such physiological functions as the induction of hypotensive, diuretic, and tranquilizing effects. It has also been implicated in the regulation of several neurological disorders, including Parkinson’s disease, Alzheimer’s disease, and Huntington’s chorea. Due to its physiological functions, the increasing commercial demand for GABA has prompted studies on the development of Health food and pharmaceutical fields [2-5]. |
| The current production of GABA methods mainly include chemical method and biological synthesis. Chemical synthesis method has the following disadvantages: chemical residues, poor safety, high cost. Microbial fermentation is a method of preparation of GABA with high efficiency and low cost. Numerous studies on GABA production from various microorganisms have been reported [6-9]. |
| GABA is primarily produced by the irreversible α- decarboxylation of acidic glutamate in a reaction catalyzed by glutamate decarboxylase (GAD). Glutamate decarboxylase (GAD) is an essential enzyme widely distributed in nature from microorganisms to plants and animals. It is the key enzyme for the biosynthesis of GABA. So the studies of scientists are still focus on the mechanism of GAD now. To find a high GAD activity of strain will become an important study for researchers [10-11]. |
| In this study, we isolated a new GABA-producing microorganism from yellow serofluid, a bean curd byproduct of production in China. This bacterial strain, LpS2, was identified as Lactobacillus plantarum. In addition, we amplified the gene of GAD from the LpS2. Then, we predicted its amino acid composition, physicochemical property, signal peptide and advanced structure by bioinformatics method. So that we can more fully understand the characteristics of GAD gene from Lactobacillus plantarum LpS2, and build a foundation for the subsequent genetic engineering bacteria. Eventually we will be able to use genetic engineering bacteria to produce GABA food and drugs by microbial fermentation method. |
Materials and Methods
|
| Strains, media and growth conditions |
| The Lactobacillus plantarum LpS2 that was found to produce a higher amount of γ-aminobutyric acid (GABA) was isolated from yellow serofluid. Centrifugal type column bacterial genome DNA extraction kitDP302-2was produced by tian gen biological engineering Ltd. Centrifugal type column DNA gel recovery kits (DP209-02) was produced by tian gen biological engineering Ltd.Trans taq hifi DNA Polymerase was produced by Shanghai treasure biological technology Ltd. |
| MRS liquid medium consists of 1% peptone, 0.5% yeast powder, 1% beef powder, 0.5% anhydrous sodium acetate, 0.2% citric acid diammonium hydrogen, 1% glucose, 0.05% MgSO4·7H2O,0.02% MnSO4, 0.2% K2HPO4, 0.1% Tween 80, pH 6.8, Liquid medium were sterilized at 121°C for 15 min before use. |
| Cloning and DNA sequencing |
| According to the inoculation quantity of 2%, the strains LpS2 were inoculated into MRS liquid medium,incubated at 37°C for 48 h. The 1.5 ml activated bacterium solution was placed in the 2 ml centrifuge tube. The cells of LpS2 were collected by centrifuging at 10000 rpm for 1min.The supernatant was discarded.180 μl lysozyme (20mg/ml)were added into cells for wall-breaking treatments and constanted temperature at 37°C for 30 min. Then 20 μl Proteinase K solution (20mg/ml) was added into solution for mixing. |
| Using the kit method for DNA extraction, solution were collected to the centrifugal tube and analyzed by 1% agarose gel electrophoresis (150 v, 100 mA) for 20 min. Then the gel of cells genomic DNA was recycled by kit method. The collected DNA was dissolved into 30μL ddH2O. |
| Lactobacillus plantarum LpS2 GAD gene was amplied by kit method. GadB-f(forward): 5'- GTAGAATTCATGGCAATGTTATACGGTAAACAC-3' and GadB-r(reverse): 5'- CTAGCGGCCGCGTGTGTGAATCCGTATTTCTTAG-3' were designed by reported Lactobacillus plantarum GAD gene sequence(Primer design was completed by sangon biological engineering co.,). The total DNA was taken as a template for amplification (Table 1). By the method of kit, Lactobacillus plantarum GAD gene PCR products were recycled. |
| The GAD gene amplification products sent to the sangon biological engineering (Shanghai) LTD for Sequencing. The sequencing results were analysised through Exploring-Local Vector NTI Database software and translated nucleotide sequence homology. Then the physical and chemical properties of protein, feeling of water analysis, protein signal peptide prediction, N-glycosylation sites prediction and protein structure domain were analysed respectively [12-15]. |
Results and Discussion
|
| PCR amplification of Lactobacillus plantarum GAD gene |
| Strain GAD gene was amplified by PCR, and then amplification products were analyzed by agarose gel electrophoresis. The result of PCR amplification of Lactobacillus plantarum GAD gene was shown in Figure 1. |
| Sequencing result of Lactobacillus plantarum GAD gene |
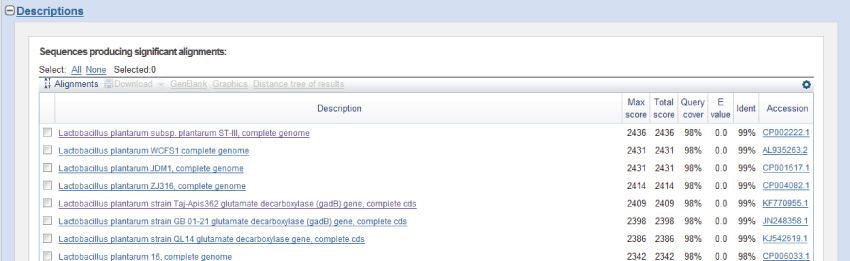
| We obtained two gene sequences by sequencing. The two gene sequences were joined together into a gene segment by software Exploring Local Vector NTI Database, which length was 1363 bp and encode 453 amino acids. By landing on NCBI (National Center of Biotechnology Information) and Using Blast program, gene segment was searched for nucleotide homology. The result was shown in Figure 2. |
| Blast analysis results showed that the amplification of gene nucleotide sequences had a very high homology with lpgadB reported in NCBI gene sequences (GenBank Accession No. KF770955.1). The deduced amino acid sequences homologies was as high as 99%.Through logged in website (http://web.expasy.org/translate/)to translation, the result showed there were a total of 453 amino acids (Figure 3). |
| Bioinformatics analysis of Lactobacillus plantarum GAD |
| By logging on website (http://web.expasy.org/protparam/) for online analysis, the formula of protein (GAD) was C2326H3541N619O663S23 and the proteins were composed of 7172 atoms. The molecular weight was 51.5 kDa and an estimated isoelectric point (pI) was 5.39. Amino acid compositions of protein (GAD) were shown in Table 2. |
| Protein hydrophilic and transmembrane region analysis of Lactobacillus plantarum GAD |
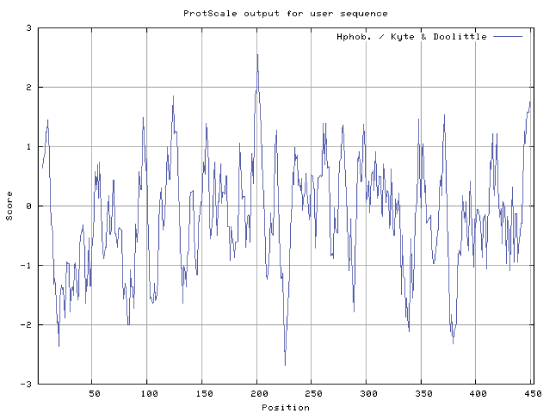
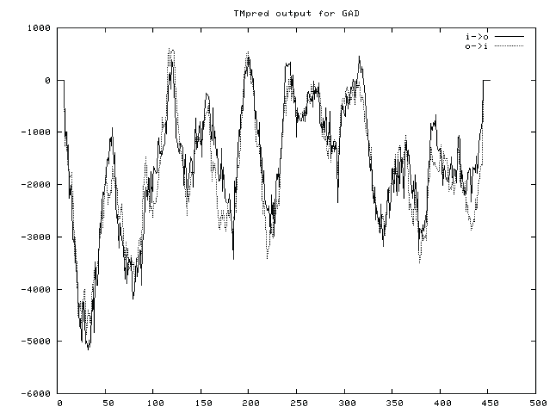
| By logging on website (http://web.expasy.org/protscale/) and website (http://www.ch.embnet.org/software/ TMPRED_form.htmL) for protein hydrophilic and transmembane region online analysis, ProtScale (a programme of ExPASy) predicted that hydrophobic amino acids GRAVY were greater than zero, and the hydrophilic amino acids GRAVY were less than zero. GRAVY of protein (GAD) was 0.179. The result indicated that the protein (GAD) was hydrophilic protein (Figures 4 and 5). |
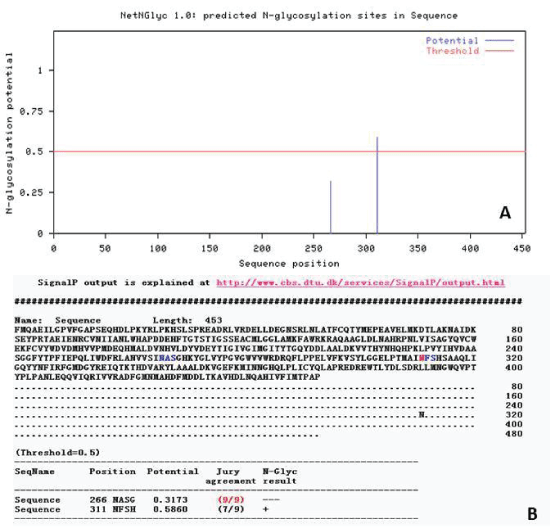
| Signal peptide prediction of Lactobacillus plantarum GAD |
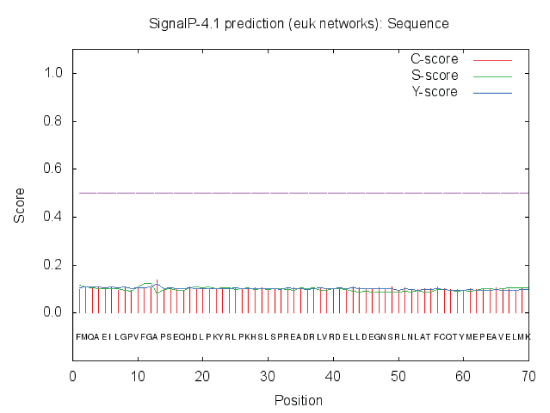
| By logging on website (http://www.cbs.dtu.dk/services/ SignalP/) for online analysis, the results of prediction were shown in Figure 6. By the value of C,S and Y value could be judged that there was no signal peptide in this protein. After the synthesis of GAD in the cytoplasm, protein was not transported. The results showed that GAD protein was not secreted proteins or some cytoplasmic matrix synthesis proteins, which was transported to organelles and played a role in there. |
| N-glycosylation analysis of Lactobacillus plantarum GAD |
| N-glycosylation is an important step of protein modification after translation. In addition, N-glycosylation has an influence on cell biology behavior of protein such as antigenicity, active, folding, transport, positioning and stability. By logging on Website (http://cbs.dtu.dk/services/NetNGlyc) to analysis and predict N - glycosylation sites, the result displayed that the Nglycosylation sites had 2 points (Figure 7). |
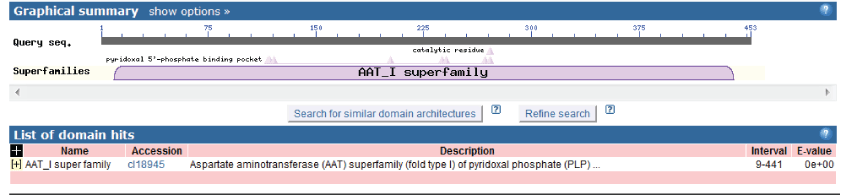
| Structure domain analysis of Lactobacillus plantarum GAD |
| By logging on website (http://www.ncbi.nlm.nih.gov/Structure/ cdd/)and using the connection in the NCBI CDD structure domain analysis, the result was shown as follow (Figure 8). Lactobacillus plantarum GAD was in the amino acid sequence 9~441 position for structural function domain. GAD gene belonged to aspartate amino transferase (AAT) superfamily (type I) pyridoxal phosphate (PLP) dependence enzymes. The comparison test results with family members (TIGR01788) were shown in Figure 9. |
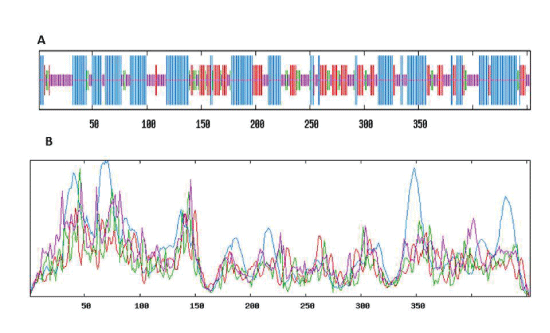
| Secondary structure prediction of Lactobacillus plantarum GAD |
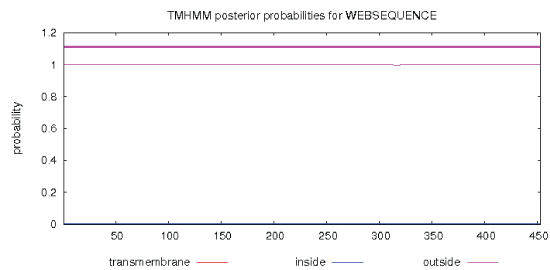
| Through logging on website (http://npsa-pbil.ibcp.fr/cgi-bin/ npsa_automat.pl?Page = npsa_sopm. HTML and http:// www.cbs.dtu.dk/services/TMHMM-2.0/), the secondary structure and transmembrane helical of GAD were analysed. The result was shown in Figure 10 and Figure 11. As shown in Figure 10, we used the SOPM online to predicte. The GAD secondary structure was composed of 43.27% alpha helix (Hh), 19.65% extension chain (Ee), 29.14% random curl(Cc) and 7.94% beta Angle (Tt). As shown in Figure 11, the protein transmembrane helical was no signal. The result indicated that the protein was not secreted proteins, and there was no transport across the membrane. |
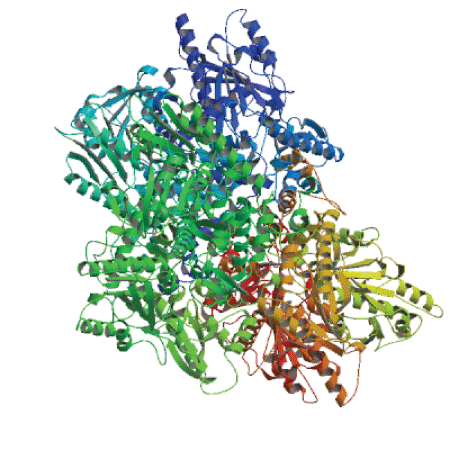
| Tertiary structure prediction of Lactobacillus plantarum GAD |
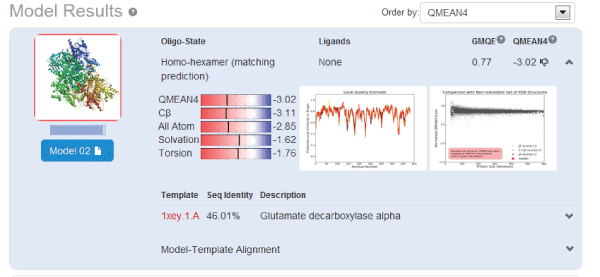
| Using homology modeling method, we logged in web site (http://swissmodel.expasy.org/) for predicting the tertiary structure of GAD gene sequence. Analysis results were shown in Figures 12 and 13. We took F chain of 1xey.1. As a template, and shaped residue cover wide range of 3-432 for modeling Blast by X-ray method. The consistency of both sequences was 46.01%. Low poly state was homologous six polymers. Resolution was 2.5A. Requence similarity was 0.41. Coverage rate was 94%. GMQE value was 0.77.QMEAN4 value was -3.02. |
| Comparison of different template on protein modeling (table 3), we took 3fz6.1.A1pmm.1.A1xey.1.A_2dgk.1.A and 3hbx.1. A as template for modeling. The coverage could reach more than 92%, and the consistency could reached more than 42%. The above showed that the gene belonged to the GAD gene. |
Conclusion
|
| In this study, a new GABA producing microorganism, Lactobacillus plantarum LpS2 was isolated from yellow serofluid.A full-length GAD gene from LpS2 was cloned. The amino acid sequence of GAD was analysised. The length of GAD gene sequence was 1363 bp and coded 453 Amino acids. GAD relative molecular weight of theoretical prediction and isoelectric point were 51521.8u and 5.39 respectively. This protein was hydrophilic protein, and there was no transmembrane region. There were two N-glycosylation sites. Structure and analysis showed that the protein belonged to the AAT super family (fold type I) pyridoxal phosphate (PLP) dependence enzyme. Secondary structure was composed of 43.27% Alpha helix, 19.65% extension chain, 29.14%random curl, 7.94% beta Angle. Transmembrane helical was no signal, and there was no transmembrane transport. The analysis of tertiary structure showed that 3fz6.1.A-1 pmm.1.A-1xey. 1.A-2dgk.1.A and 3hbx.1. A were taken as template for modeling, and the coverage could reach more than 92%, and the consistency could reached more than 42%. |
| This research adopted the bioinformatics analysis method, which was considered to be authoritative in the biological field. This result was very important for the expression, separation and purification of glutamate decarboxylase in the following work. In addition, it is very meaningful for producting GABA food and drug by genetic engineering strains. |
Acknowledgment
|
| This work was supported by Heilongjiang University of Science and Technology innovation team building program, PR China. |
Tables at a glance
|
|
|
Figures at a glance
|
 |
 |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
Figure 4 |
Figure 5 |
 |
 |
 |
 |
 |
| Figure 6 |
Figure 7 |
Figure 8 |
Figure 9 |
Figure 10 |
 |
 |
 |
| Figure 11 |
Figure 12 |
Figure 13 |
|
References
|
- Manyam BV, Katz L, Hare TA, Kaniefski K, Tremblay RD. Ann Neurol, 1981; 10: 35-37.
- Hayakawa K, Kimura M, Kasaha K, Matsumoto K, Sansawa H, Yamori Y. Effect of a gamma-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar-Kyoto rats. Br J Nutr 2004; 92:411-417.
- Inoue K, Shirai T, Ochiai H, Kasao M, Hayakawa K, Kimura M, Sansawa H. Blood-pressure-lowering effect of a novel fermented milk containing gamma-aminobutyric acid (GABA) in mild hypertensives. Eur J ClinNutr 57: 490-495.
- Jakobs C, Jaeken J, Gibson KM. Inherited disorders of GABA metabolism. J Inherit Metab Dis 1993; 16:704-715.
- Wong CG, Bottiglieri T, Snead OC. GABA, gamma-hydroxybutyricacid and neurological disease. Ann Neurol 2003; 54:S3-12.
- Nomura M, Nakajima I, Fujita Y, Kobayashi M, Kimoto H, Suzuki I, Aso H.Lactococcuslactis contains only one glutamate decarboxylase gene.Microbiology1999; 145: 1375-1380.
- Park KB, Oh SH. Cloning,sequencing and expression of a novel glutamate decarboxylase gene from a newly isolated lactic acid bacterium, Lactobacillus brevis OPK-3. Bioresource Technology2007; 98: 312-319.
- Park KB,Heung OS. Cloning and expression of a full-length glutamate decarboxylase gene from Lactobacillus plantarum.Journal of Food Science and Nutrition2004; 9: 324-329.
- Kim SH, Shin BH, Kim YH. Cloning and expression of a full-length glutamate decarboxylase gene from Lactobacillus brevis BH2.Biotechnology and Bioprocess Engineering2007; 12: 707-712.
- Naidu AS, Bidlack WR, Clemens RA. Probiotic spectra of lactic acid bacteria (LAB). Crit Rev Food SciNutr 1999; 38: 13-126.
- Aymerich MT, Garriga M, Monfort JM, Nes I, Hugas M. Bacteriocin-producing Lactobacilli in Spanish-style fermented sausages: characterisationof bacteriocins. Food Microbiol 2000; 17: 33-45.
- Leenhouts KJ,Kok J,Venema G. Stability of Integrated Plasmids in the Chromosome of Lactococcuslactis. Appl EnvironMicrobiol1990; 56: 2726-2735.
- de Jong A,van Heel A J,Kok J,et al.BAGEL2: mining for bacteriocins in genomic data.Nucleic Acids Res 2010; 38: W647-W651.
- Maldonado A,Ruiz-Barba JL,Jimenez-Diaz R. Purification and genetic characterization of plantaricin NC8,a novel coculture -inducible two - peptide bacteriocin from Lactobacillus plantarum NC8.Appl Environ Microbiol2003; 69:383-389.
- Rojo-Bezares B, Sáenz Y, Navarro L, Zarazaga M, Ruiz-Larrea F, Torres C.Coculture-inducible bacteriocin activity of Lactobacillus plantarum strain J23 isolated from grape must.Food Microbiol2007; 24: 482-491.
|












