Research Article - Biomedical Research (2016) Computational Life Sciences and Smarter Technological Advancement
Classification of mammogram masses using selected texture, shape and margin features with multilayer perceptron classifier
P Valarmathie1*, V Sivakrithika2 and K Dinakaran21Department of Computer Science and Engineering, Saveetha Engineering College, Kanchipuram, Tamil Nadu, India
2Department of Computer Science and Engineering, PMR Engineering College, Chennai, Tamil Nadu, India
- *Corresponding Author:
- P Valarmathie
Department of Computer Science and Engineering
Saveetha Engineering College
Tamil Nadu
India
Accepted date: September 26, 2016
Abstract
Computer-aided detection (CAD) assists radiologists by providing the second opinion in the mammography detection and reduces misdiagnosis. In this work the task of automatically classifying the mass tissue into benign and malign based on the characteristics of mass is investigated. Mass is characterized by its shape, margin, density, size and age of the patient. Geometrical shape, margin and texture features are used in this work to classify the masses. These features are found to be effective in discriminating benign mass from the malign mass. For the purpose of classification, the masses are segmented from the mammogram using gray level thresholding and features are extracted. Then the features are fuzzified using fuzzy membership values. Finally, the classification is performed using different classifiers and their performances are compared. Mammographic Image Analysis Society (MIAS) Database was used for experimental study. The experiments were implemented in MATLAB and WEKA.
Keywords
Medical image analysis, Data mining, Cancer diagnosis, Mammogram classification, Computer aided diagnosis, Feature extraction, Feature selection, Classifiers, Mammogram abnormalities.
Introduction
Breast cancer is the second most common type of cancer in the world, more common among women and representing 22% of all new cancer cases every year. The earlier it is diagnosed, the better the chances of a successful cure. Mammography [1] is the best, cheapest way to detect tumors that cause breast cancer with fewer side effects. However, it is known that the sensitivity of this examination can vary considerably due to factors such as the specialist's experience, human error and the quality of the images obtained in the exam. The use of computational techniques involving machine learning and image processing has contributed more and more to support the radiologists as the second reader in procuring a more precise report.
Mammography is considered as the successful screening method for the detection of abnormalities present in the breast. However, it is known that even skilled radiologists can miss a considerable amount of abnormalities due to overload. In most cases after biopsy, a large number of mammographic abnormalities end up being benign, this may cause unnecessary discomfort to the patient. There are five abnormalities in the mammograms that are suspicious which requires further examination to classify it as benign or malign. The abnormalities are:
1. Asymmetric breast tissue
2. Mass
3. Micro calcification
4. Asymmetric density
5. Architectural distortion
Among the five abnormalities except for mass and micro calcification all the other abnormalities are found in very rare cases known as invasive breast cancer of low risk. It is found that only 5-7% of women represent these rare case. Mass and micro calcification are the two most suspicious abnormal sign of breast cancer. It is accepted that mass classification as benign and malign is harder and challenging problem.
Mass is a space occupying lesion Mass is the cluster of cells denser than the surrounding tissue. The mass can be identified using the following subtle visual clues such as shape, margin, and density. In addition to that patient age and history also plays a vital role in diagnosing process. It is stated from several studies that the CAD improves breast cancer diagnostic accuracy rate by 14.2% and reduces misinterpretation rate. There exist impreciseness and ambiguity in describing the mass in mammogram using features, so the feature values extracted are fuzzified. In this paper, for each mass, shape, density and margin properties are extracted as feature vector, and membership function is used to discretized.
The rest of the paper is organized as follows. Section 2 depicts the related works in mammogram classification. The proposed methodology is illustrated in Section 3. Feature Extraction and Fuzzification are discussed in Section 4 and 5.Rest of the paper shows how the classifier performance is evaluated.
Related Work
In [2], thresholding segmentation had been used, where pixels with gray level values greater than the defined threshold are retained while all others are set to zero. Then statistical texture features are extracted from this ROI. Breast masses appear in the dense regions in mammograms. Benign masses are round or regular, smooth with well-circumscribed boundaries and have homogeneous texture. And malign mass are irregular, rough, spiculated or blurry boundaries and heterogeneous texture [3]. Several shape features have been proposed for the classification of benign masses and malignant tumors. The different feature set representing radiological characteristics of the masses such as shape, edge-sharpness, and texture features have been evaluated using several pattern classification methods individually [4-6] including several linear classifiers, artificial neural networks (ANNs), and kernel-based classification methods. The most common mass detection feature extraction technique used in literature is Gray-Level Co-occurrence Matrix (GLCM). In [7,8] five co-occurrence matrices statistics extracted from four spatial orientations, and pixel distance (d=1). The extracted features are not discriminative with cases of cancers due to their non-uniform shape and margins.
Vadivel et al. [9] classified mass lesion into BIRADS shape categories using geometric shape and margin features and C5.0 decision tree classifier with fuzzy inference system. Among the various categories, classifying irregular and oval shape mass had the highest performance with 97% accuracy. The features used are Elongatedness, Mass edge Standard deviation, Max radius, Shape index, Standard deviation of mass, Dispersion. In artificial neural network is used to classify masses in mammograms as malignant or benign incorporating three shape factors, three measures of edge sharpness, and 14 texture features based on gray-level co-occurrence matrices of the pixels in the ribbons are computed [10]. The feature sets that composed of shape factor provide high performance with respect to different network structure and learning rule. In a set of 22 features including five shape factors, three edgesharpness measures, and 14 texture features computed and SVM, Fishers LDA classifier and their nonlinear versions are used. ROC of 0.95 has been achieved [11].
Methodology
In this work, the first step involves the pre-processing stage in which labels and artifacts present in the mammogram are removed, and the median filter is used to denoise the image. As a second step region of interest is segmented using thresholding technique. To diagnose a mass in mammogram significant features that discriminate malignant from benign mass based on the visual features such as shape, size, margin and texture are used on radiologist’s opinion and experience. As third step features are extracted and the values are fuzzified. Finally, different classifiers are used, and their performance is evaluated.
Feature extraction
The third stage of mass detection by CAD (computer aided diagnosis) schemes is the feature extraction and selection. The features can be calculated from the ROI characteristics such as the size, shape, density, smoothness of borders. It is imperative from the study made that combination of features from different perspectives is necessary. So in the proposed model selected shape, margin, texture features are extracted from the image. Along with these features size of the mass and patient age is also considered.
Shape features
According to the radiologist characteristics of mass, shape properties discriminate the benign and malign class to the most. In this work along with standard morphological features other features such as compactness, shape index and dispersion are used. Table 1 shows the shape feature used in this work.
| Feature | Formulae | Description |
|---|---|---|
| Compactness |  |
Circle takes value of 1, while other irregular shapes have low value |
| Dispersion |  |
Identifies irregular shape characteristics |
| Eccentricity | 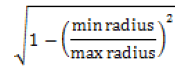 |
0 for circle and maximum value for other shapes |
| Elongatedness | 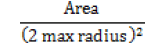 |
For circle and oval closer to 1 and other shapes closer to 0 |
| Roundedness |  |
Higher value for circle and lower value for other shapes |
Table 1. Shape features discriminating mass patterns.
Margin features
Margin features depend on boundary descriptors of mass. For an irregularly shaped object, the boundary descriptor is the best way of representation as they are not used in shape features. In this work we have made use of the following margin features as shown in Table 2.
| Feature | Formulae | Descriptor |
|---|---|---|
| Convexity | 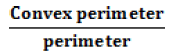 |
1 for convex shape and lower for the object whose perimeter is rough |
| Standard deviation of edge | 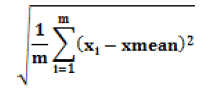 |
Average contrast of the mass boundary large deviation for larger dispersion |
| Shape Index | 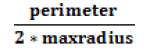 |
Surface curvature |
Table 2. Margin features discriminating.
Texture features
Texture features play a vital role in medical image analysis. In this work, we have incorporated texture features based on intensity histogram features and Grey Level Co-occurrence Matrix (GLCM) as shown in the Tables 3 and 4.
| Feature | Formulae | Description |
|---|---|---|
| Average Intensity | 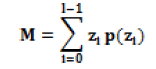 |
Average Intensity (More for dense tissue) |
| Standard deviation | 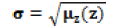 |
Average contrast(Irregularity of the texture) |
| Smoothness | 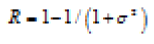 |
Smoothness of the intensity in the region(low to regular intensity level ie smooth) |
| Uniformity | 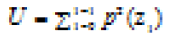 |
Uniformity of intensity in the histogram(high in soft tissues) |
| Third Moment | 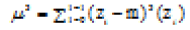 |
Skewness of the histogram |
Table 3. Histogram based intensity features discriminating mass patterns.
| Feature | Formulae | Description |
|---|---|---|
| Correlation | 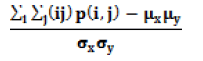 |
A measure of gray tone linear dependencies in the image. |
| Sum variance | 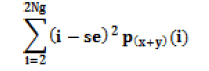 |
Contrast of the image |
| Information Measure of Correlation1 | 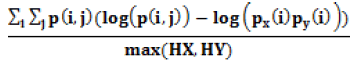 |
Correlation of a pixel is to its neighborhood |
| Information Measure of Correlation2 | 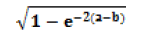 |
|
| Sum Entropy | 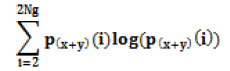 |
Randomness |
| Energy | 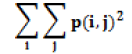 |
Homogeneity of the image |
Table 4. GLCM based texture features.
Fuzzy discretization
The crisp data set leads to loss of information at the boundaries of ranges. The proposed work makes use of fuzzy partition method. For each feature trapezoidal fuzzy membership function is calculated using the formula
α=2β-ϒ Where α=lower end range
β=average value of the feature fi in the database
where f is the feature and i=1 to 15
ϒ=the largest value of the feature fi in the database
The probability of the crisp dataset value with respect to the membership function is known as fuzzy categorical value of the features. More over fuzzy discretization technique [12] provides high accuracy while classifying the test images.
Mammogram classification
Classification of mammogram automatically is important for Radiologists to reduce the misdiagnosis rate. In this work, several features are extracted from the mammographic masses and given as the input to the classifier. It has been proved that Neural Network and SVM classifier outperformed others in mammogram classification. In this work, we have used MultilayerPerceptron and Radial Basis Function.
Experimental Results
The performance of the classifier and the combination of features are evaluated using MATLAB and WEKA. To show the difference between classification accuracy on the combination of feature characterizing the mammogram mass, we have shown the comparison between them on the same dataset. The mini-MIAS database is used for classification. The classification has been made using the cross-validation with ten-fold. As the Mini MIAS database consists of 332 mammograms, it has been selected for the testing of the performance of the proposed work. Among that 200 images are used for training purpose and 132 images for testing purpose. Each feature sets are individually and their combinations are used to evaluate the performance of the classifier. The results obtained are tabulated in the table. From the experimental results, it can be clearly seen that the combinations of features play a vital role in the performance of the classifier. MLP classifier with all the three feature set outperformed with an accuracy of 100% and 0.99 ROC curve. In both the classifier combination of margin and shape feature has also shown greater accuracy.
Conclusion
The mammographic masses were classified into benign or malignant. It is found that the masses, which are represented using shape and margin properties, have certain amount of impreciseness to overcome this fuzzification is done. The experimental results from Table 5 and Figure 1 indicate that the combination of texture, shape and margin features are effective in classifying the mammographic masses. It is also found from the results that combination of shape and margin feature also gives good accuracy.
| Classifier | Feature Set | ROC | Accuracy |
|---|---|---|---|
| MLP | Texture Features | 0.774 | 0.68 |
| MLP | Shape Features | 0.861 | 0.85 |
| MLP | Margin Features | 0.642 | 0.67 |
| MLP | Texture and Shape Features | 0.937 | 0.851 |
| MLP | Texture and Margin Features | 0.934 | 0.8235 |
| MLP | Margin and Shape features | 0.94 | 0.93058 |
| MLP | Texture, Shape and Margin Features | 0.96 | 0.98 |
| RBF | Texture Features | 0.76 | 0.62 |
| RBF | Shape Features | 0.917 | 0.82 |
| RBF | Margin Features | 0.799 | 0.735 |
| RBF | Texture and Shape Features | 0.875 | 0.823 |
| RBF | Texture and Margin Features | 0.806 | 0.705 |
| RBF | Margin and Shape features | 0.938 | 0.882 |
| RBF | Texture, Shape and Margin Features | 0.872 | 0.8235 |
Table 5. Classifier accuracy based on feature group combinations.
References
- Homer MJ. Mammographic Interpretation: A Practical Approach. 2. Boston: McGraw-Hill; 1997.
- Subashini TS, Ramalingam V, Palanivel S. Automated assessment of breast tissue density in digital mammograms. Compute Vision Image Understanding 2010; 114: 33-43.
- Bovis K, Singh S. Detection of masses in mammograms using texture features. 15th International Conference on Pattern Recognition (ICPR'00), 2000.
- Sahiner BS, Chan HP, Petrick N, Helvie MA, Hadjiiski LM. Improvement of mammographic mass characterization using spiculation measures and morphological features. Med Phys 2001; 28: 1455-1465.
- Rangayyan RM, El-Faramawy NM, Desautels JEL, Alim OA. Measures of acutance and shape for classification of breast tumors. IEEE Trans Med Imaging. 1997; 16: 799-810.
- Nandi RJ, Nandi AK, Rangayyan RM, Scutt D. Classification of breast masses in mammograms using genetic programming and feature selection. Med Biol Eng Comput 2006; 44: 693-694.
- Wei D. Classification of mass and normal breast tissue on digital mammograms: multiresolution texture analysis. Phys Med 1995; 22: 1501-1513.
- Surendiran B, Vadivel A. Feature Selection using Stepwise ANOVA Discriminant Analysis for Mammogram Mass Classification. Int J Recent Trend Eng Technol 2010; 3: 55-57.
- Surendiran B, Vadivel A. Mammogram mass classification using various geometric shape and margin features for early detection of breast cancer. Int J Med Eng Informa 2012; 4: 36-54.
- Túlio CSSA, Rangaraj MR. Classification of breast masses in mammograms using neural networks with shape, edge sharpness, and texture feature . J Elect Imaging 2006.
- Mu T, Nandi AK, Rangayyan RM. Classification of Breast Masses Using Selected Shape, Edge-sharpness, and Texture Features with Linear and Kernel-based Classifiers J Digit Imaging 2008; 21: 153-169.
- Sivakrithika V, Shanthi B. Comparative Study on Cancer Image Diagnosis using Soft Computing Techniques. Int J Comput Appl 2011.
