Review Article - Journal of Biochemistry and Biotechnology (2025) Volume 8, Issue 1
Characterization, Partial purification and Optimization of Cellulase enzyme production from bacteria isolated from mangrove soil of Bhitarkanika, Odisha
Swetlina Sahoo*, Amisha Biswal, Priyanka PrustyDepartment of Biotechnology, MITS School of Biotechnology, India
- *Corresponding Author:
- Swetlina Sahoo
Department of Biotechnology
MITS School of Biotechnology, India
E-mail: swetlinasahoo1609@gmail.com
Received: 10-Feb-2025, Manuscript No. AABB-25-161089 Editor assigned: 11-Feb-2025, Pre QC No. AABB-25-161089 (PQ); Reviewed: 24-Feb-2025, QC No. AABB-25-161089; Revised: 26-Feb-2025, Manuscript No. AABB-25-161089 (R); Published: 28-Feb-2025, DOI:10.35841/aabb-8.1.241
Citation: Sahoo S. Characterization, Partial purification and Optimization of Cellulase enzyme production from bacteria isolated from mangrove soil of Bhitarkanika, Odisha. J Biochem Biotech 2025; 8(1):241.
Abstract
The mangroves of Bhitarkanika, Odisha, harbor a diverse microbial population, including cellulose-degrading, phosphate-solubilizing, sulfur-oxidizing, and reducing bacteria. Among these, cellulose-degrading bacteria are dominant due to the nutrient-rich soil. Cellulases are in high demand for converting lignocellulosic materials into renewable energy, chemicals, and biofuels. This study aimed to isolate cellulolytic bacteria from Bhitarkanika mangrove soil and assess their cellulase production using CMC agar. Out of 20 bacterial isolates, 10 demonstrated cellulase activity, ranging from 0.014±0.004 to 0.267±0.007. The isolate BCDB10, showing high cellulolytic activity, was optimized for enzyme production using response surface methodology (RSM). The enzyme was purified through ammonium sulfate precipitation and DEAE-cellulose chromatography, revealing a molecular mass of 54 kDa. Phenotypic characterization identified BCDB10 as a Gram-positive, non-motile, spore-forming rod, presumptively Bacillus. While BCDB10 exhibited significant cellulase activity, further studies are needed to evaluate its industrial potential for cellulase production.
Keywords
Cellulose, Morphology, Bacteria, Enzyme, Biological, Microbial.
Introduction
Bhitarkanika mangroves in Odisha, located at the confluence of the Brahmani and Baitarani rivers, form the second-largest mangrove ecosystem in India, after the Sundarbans in West Bengal. This ecosystem is globally recognized for its rich biodiversity, ranking among the world's top mangrove forests. The biological diversity in Bhitarkanika is extraordinary, with a variety of plant and animal species. However, this valuable ecosystem is under threat due to deforestation and developmental activities, leading to a steady decline in mangrove vegetation.
While several studies have focused on the flora and fauna of Bhitarkanika's mangroves, the microbial diversity of this habitat has largely been overlooked. Microbes play a crucial role in the biogeochemical cycles that sustain these ecosystems, yet little is known about their diversity and functions within Indian mangrove systems. Understanding the microbial diversity, particularly cellulase-producing bacteria, is essential for understanding nutrient cycling and biotechnological applications [1].
In this study, an attempt was made to isolate and characterize cellulase-producing bacteria from the Bhitarkanika mangrove ecosystem and optimize their enzyme production. Cellulases are of significant industrial interest due to their ability to convert lignocellulosic materials into renewable energy, chemicals, and biofuels. The study aimed to explore the cellulolytic potential of bacterial isolates from mangrove soil and optimize their cellulase production using different methodologies [2]. This investigation addresses the gap in microbial diversity studies within the Bhitarkanika ecosystem, offering insights into the biogeochemical processes of this unique habitat and highlighting the potential industrial applications of cellulase-producing bacteria from mangrove soils.
Importance of microbial cellulase enzyme
Over the past two decades, cellulase has gained significant attention as an industrial enzyme with a wide range of applications. It plays a key role in the saccharification of cellulose, the main component of lignocellulosic biomass, releasing glucose that can be converted into biofuels like bioethanol using ethanologenic microorganisms. Cellulase, along with pectinase, is also used in food processing for fruit juice extraction and clarification. Currently, cellulase ranks as the third most important industrial enzyme globally. The production of cellulase is crucial for the economic conversion of renewable lignocellulosic materials into value-added products like ethanol, single-cell proteins, and chemicals. However, its production is expensive, contributing to 50% of total hydrolysis costs. Exploring diverse ecological habitats for new microbial cellulase producers is essential to meet the growing demand for this enzyme.
Classification and mechanism of action of cellulolytic enzymes
Cellulolytic enzymes are categorized into three main groups: endoglucanases (EC 3.2.1.4), exoglucanases (cellobiohydrolase, EC 3.2.1.91, and cellodextrinase, EC 3.2.1.74), and β-glucosidases (EC 3.2.1.21). These enzymes belong to distinct glycoside hydrolase (GH) families as classified by the CAZy database. The classification of cellulases is based on their depolymerization stage of the substrate. Endoglucanases hydrolyze glycosidic bonds in both crystalline and amorphous cellulose, producing oligomers with varying degrees of polymerization. Studies showed that endoglucanases are more effective on crystalline cellulose, while amorphous cellulose is more susceptible to exoglucanases, which hydrolyze β-1,4-glycosidic bonds to produce cellobiose. This cellobiose is further degraded to glucose by β-glucosidases [3].
Additionally, enzymes like cellobiose phosphorylase and cellodextrin phosphorylase are involved in the reversible phosphorolysis of cellobiose and cellodextrins, converting them to glucose. The cellulase enzyme complex also includes accessory proteins such as swollenin and lytic polysaccharide monooxygenases, which assist in cellulose degradation.
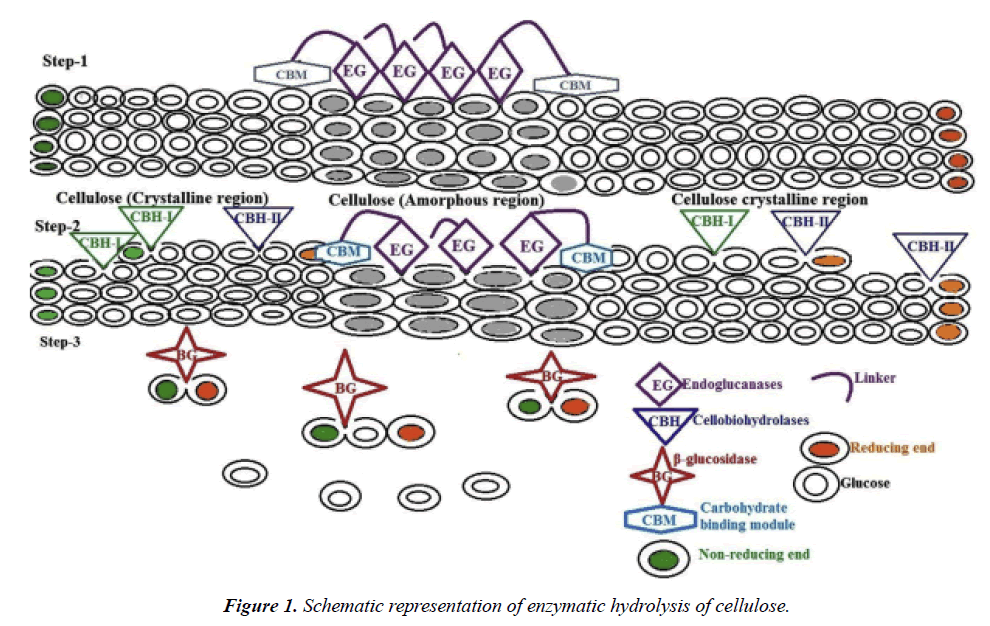
Trichoderma reesei, a widely used industrial cellulase producer, produces a range of enzymes, including cellobiohydrolases, endo-1,4-β-glucanases, β-glucosidase, and lytic polysaccharide monooxygenase, essential for effective cellulose degradation. The enzymatic saccharification rate improves when crystalline cellulose I is disrupted into other crystalline forms. The synergistic action of the cellulase enzyme complex involves two steps: first, the exo- and endoglucanases reduce the polymerization degree of cellulose, releasing cellobiose, and second, β-glucosidase converts cellobiose into glucose [4]. The cellulase adsorption on cellulose is crucial for efficient hydrolysis, and the cellulose-binding domain plays a key role in this process (Figure 1).
Cellulose degrading bacteria in mangrove ecosystem
The search for potential cellulolytic enzymes continues to be a focal point for the successful bioconversion of lignocellulosic biomass, which is crucial for bioenergy production. Numerous studies have highlighted the rich microbial diversity in mangrove ecosystems around the world, particularly in the context of cellulose degradation. These microbial communities, including bacteria, fungi, yeast, and actinomycetes, play a key role in breaking down the complex lignocellulosic materials found in mangrove habitats.
Several reports have documented bacterial isolates capable of degrading cellulose from mangrove environments. He isolated cellulose-degrading bacteria from decaying Spartina alterniflora plants in salt marshes of Sapelo Island, Georgia. He reported a wide variety of cellulose-degrading bacteria from the Sundarban mangroves of West Bengal, India. Similarly, they isolated five cellulase-producing strains, including Bacillus cereus, Bacillus licheniformis, Bacillus pumilus, and Bacillus sp., from Philippine mangroves. He identified a cellulolytic bacterium, Vibrio xiamenensis, isolated from mangrove soil in Xiamen, China. In Odisha, India, He isolated seven cellulose-degrading bacterial species from Bhitarkanika mangrove soil, including Pseudomonas sp.., Bacillus polymyxa, Bacillus mycoides, and Bacillus brevis. Bacterial species with endo- and exoglucanase activities were also reported in the sediments of a Brazilian mangrove. He isolated two cellulolytic bacteria, Paenibacillus sp. and Bacillus sp., from the Sundarban mangrove [5].
In addition to bacteria, fungi have also been recognized for their cellulolytic potential in mangrove ecosystems. He reported several cellulose-degrading fungi from Avicennia marina roots along the Red Sea Coast of Egypt, including Aspergillus niger, Cladosporium cladosporioides, Penicillium chrysogenum, and Stachybotrys chartarum. He screened twenty-nine fungal isolates from mangrove environments in Thailand, Hong Kong, and Vietnam, finding that most of them were ascomycetes, with significant endoglucanase production. In India, He identified seven fungal species from the Nethravathi mangrove, including Acremonium sp., Alternaria sp., Aspergillus sp., and Fusarium sp.,that exhibited cellulase enzyme activity. Other significant reports include who isolated Penicillium fellutanum from coastal mangrove soil in India, who found that Aspergillus niger from Indian mangrove soil produced the highest cellulase activity when grown on wheat bran [6].
Actinomycetes have also been reported to degrade cellulose in mangrove ecosystems. They identified actinomycetes with cellulase activity from the Konkan Coast of Maharashtra, including Streptomyces sp., Micromonospora sp., Rhodococcus sp., and Nocardia sp.. In Odisha, He isolated nine cellulose-degrading actinomycetes from Bhitarkanika mangrove soil, while he identified 58 cellulase-producing actinomycetes from the rhizosphere of mangrove plants along the Jazan Coast in Saudi Arabia. Actinomycetes from the Coringa mangrove forest in Andhra Pradesh, India, such as Streptomyces sp., were also reported to produce cellulose [7].
In addition to bacteria, fungi, and actinomycetes, other microorganisms like yeast, such as Pichia salicaria, Cryptococcus dimennae, and Pichia fermentans, have been isolated from decomposing leaves of Rhizophora mucronata and Avicennia marina in the Vellar estuary mangrove forest of India. The high diversity of cellulolytic microorganisms in mangrove environments underscores the potential of these ecosystems as rich sources of cellulase-producing microbes, which can be harnessed for industrial applications, particularly in bioconversion processes.
Mechanism of cellulose hydrolysis by cellulase enzyme
Cellulases are enzymes that break down cellulose by hydrolyzing the β-1,4-glycosidic bonds in the cellulose polymer. Complete hydrolysis of cellulose into glucose requires the synergistic action of three main enzymes: endoglucanases (EG), cellobiohydrolases (CBH), and β-glucosidases (BG). Endoglucanases, which primarily attack the amorphous regions of cellulose, randomly cleave internal glycosidic bonds, creating cellooligosaccharides with reducing or non-reducing ends. These products then provide substrates for cellobiohydrolases, which cleave the chain ends in a processive manner, producing cellobiose as the main product. Finally, β-glucosidases hydrolyze cellobiose into glucose and release glucose from the non-reducing ends of soluble cellooligosaccharides [8].
The action of CBH and EG enzymes is highly synergistic, ensuring efficient cellulose degradation. The products of their activities—cellodextrins and cellobiose—are inhibitory to enzyme function. Thus, the presence of β-glucosidases is essential to prevent these inhibitory effects by converting the final products into glucose. Some bacteria also produce intracellular or extracellular β-glucosidases to further cleave cellodextrins and cellobiose, releasing glucose for assimilation by the cell.
Mechanistically, cellulases utilize acid-base catalysis, where a carboxylate pair at the enzyme's active site helps break the glycosidic bond. Depending on the distance between the carboxylate groups, cellulases exhibit either inverting or retaining mechanisms. Unlike soluble substrates, cellulose is insoluble and requires cellulases to bind and move along the cellulose polymer to access their active sites. Most cellulases are modular proteins, with catalytic modules connected by a flexible linker to one or more carbohydrate-binding modules (CBMs) [9]. CBMs help bind the enzyme to cellulose, enhancing its activity, and some are also capable of disrupting crystalline cellulose, making cellulase activity more effective against insoluble polysaccharides.
Overview of cellulolytic enzymes production
Microorganisms, particularly bacteria and fungi, are excellent producers of cellulolytic enzymes, with fungi being preferred due to their extracellular enzyme production. Ongoing exploration for new microorganisms is crucial to meet industrial demands. Commercially, cellulases are produced using substrates like cellulose or carboxymethyl cellulose (CMC), but these are costly. To reduce production costs, researchers have focused on using cellulose-rich biomass from agricultural and forest residues as an alternative substrate for cellulase production.
Utilization of pure cellulose as a substrate and suitable carbon source for cellulase production
Pure cellulose derived from microbial and biomass processing can serve as a substrate for cellulase production. He demonstrated the use of commercially available cellulose, such as carboxymethyl cellulose (CMC), for cellulase production by the fungal strain Schizophyllum commune NAIMCC-F-03379. They compared CMC with various lignocellulosic biomass (LCB) substrates and found that wheat bran and CMC exhibited comparable CMCase and FPase activities. In contrast, other biomass sources, including rice straw, rice husk, wheat straw, and sugarcane bagasse, showed lower cellulase activity [10]. While fungi are commonly used for cellulase production, cellulase-producing bacteria have gained significant attention due to their strong adaptability. For instance, Bacillus velezensis was reported to have the highest enzyme production ability among ten cellulase-producing bacterial strains isolated from pig manure.
Cellulase production using Whatman filter paper rich in cellulose as a substrate has also been demonstrated, where cellulolytic bacteria were used to produce enzymes for simultaneous saccharification and fermentation (SSF), leading to efficient ethanol production. Additionally, recombinant expression of thermostable cellobiohydrolase from Chaetomium thermophilum in Pichia pastoris showed high saccharification efficiency with cellulose-rich substrates, demonstrating thermotolerant and acid-stable enzyme properties [11].
The carbon source used in enzyme production can constitute over 50% of the total cost, such as pure glucose. To reduce costs and ensure sustainability, researchers are exploring lignocellulosic biomass from agricultural and forestry residues, which are abundant and cost-effective. These residues, including wheat bran, sugarcane bagasse, rice straw, and fruit pomace, provide a renewable and nutrient-rich alternative for cellulase production [12]. Sugarcane bagasse, in particular, is a viable option due to its large availability in Brazil’s sugar mills. Lignocellulosic residues not only reduce production costs but also act as enzyme inducers, encouraging the production of enzymatic cocktails capable of efficiently breaking down cellulose.
Media optimization and process parameters using statistical tools for enhanced cellulase production
Various optimization studies have been conducted to enhance cellulase production, using methods such as One Factor at a Time (OFAT) and statistical approaches. These studies demonstrated that optimizing medium components and physicochemical conditions significantly improved enzyme yield. A widely used statistical tool, Response Surface Methodology (RSM), has been applied to optimize cellulase production by evaluating the interactions of independent physicochemical parameters in strains like A. aneurinilyticus BKT-9 and Schizophyllum commune COC. RSM offers advantages such as fewer experimental runs and better understanding of the effects of individual and interactive parameters. The Central Composite Design (CCD), a key component of RSM, helps optimize nutritional and environmental factors for cellulase production [13]. He optimized cellulase production by Bacillus licheniformis KY962963 using Plackett-Burman design and OFAT, identifying key factors like moisture content, K2HPO4 concentration, and temperature.
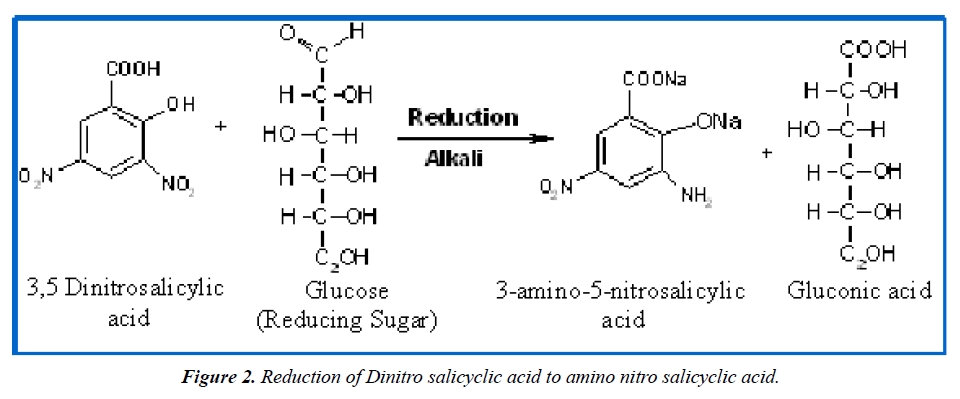
Principle of Calorimetric Determination of cellulase by the 3,5-dinitrosalicylic acid Method
Several reagents assay sugars by their reducing properties, detecting free carbonyl groups in reducing sugars like glucose and fructose [14]. This process oxidizes the aldehyde and ketone groups while reducing 3,5-dinitrosalicylic acid to 3-amino-5-nitrosalicylic acid (Figure 2).
Materials and Methods
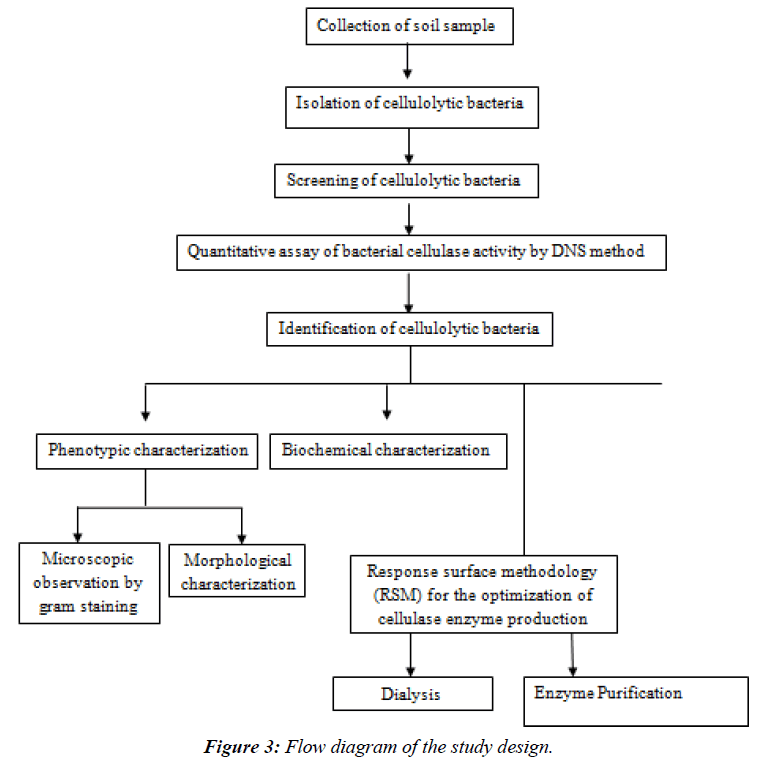
The flow chart for the methods is as follows: (Figure 3)
Collection of Soil Samples
Soil samples for this study were collected from five different locations within the mangrove forest of Bhitarkanika, Odisha. The top 1 cm of soil was removed using a sterile spatula, and the remaining soil was collected in sterile polythene bags. After collection, the samples were transported immediately to the MITS laboratory and stored at 4°C for further analysis.
Isolation of Cellulolytic Bacteria
Serial Dilution Method
One gram of each soil sample was homogenized in 9 ml of sterilized double-distilled water (dd H2O) in a test tube. The suspension was serially diluted to 10^-5 and 10^-6 concentrations.
Pour Plate Method
To isolate different bacterial sub-colonies from the serially diluted soil samples, the pour plate method was used. The CMC agar medium was prepared with the following composition (g/L): Carboxymethylcellulose (CMC), 26g; KH2PO4, 4g; K2HPO4, 1g; MgSO4·7H2O, 2g; KCl, 0.2g; yeast extract, 1g; Agar, 3g. After inoculating the plates with the serially diluted soil samples, the CMC agar (cooled to 40°C) was poured onto the plates and gently swirled in an "8" shape to ensure even distribution [15]. The plates were allowed to set for 10-15 minutes before sealing them with parafilm and incubating at 37°C for 24-48 hours.
Preparation of Pure Bacterial Isolates
To further analyze enzymatic activity and perform phenotypic and molecular characterization, pure bacterial isolates were obtained through the streak plate method.
Streak Plate Method
Individual bacterial sub-colonies from the pour plates were selected based on their morphological characteristics, including shape, size, and color. The streak plate medium consisted of 13 g of nutrient broth and 25 g of agar per liter. Using a sterile inoculating loop, bacterial colonies were transferred to nutrient agar plates and incubated at 37°C for 24 hours. The process was repeated until pure bacterial cultures were obtained.
Preservation of Bacterial Isolates
After isolating pure bacterial cultures, their viability and purity were maintained through proper preservation methods.
Bacterial Culture in Nutrient Agar Slants
Pure bacterial isolates were inoculated onto 5 ml of nutrient agar (nutrient broth, 0.26 g; agar, 0.5 g). The slants were sealed with non-absorbent cotton and parafilm to prevent contamination and incubated at 37°C for 24 hours. The slants were then stored at 4°C for long-term preservation [16].
Bacterial Culture in Nutrient Broth
Bacterial isolates from nutrient agar slants were transferred to 5 ml of nutrient broth and incubated at 37°C for 24 hours. After growth, the broth became turbid and was stored at 4°C for further use.
Screening of Cellulolytic Bacteria
The cellulolytic activity of bacterial isolates was screened using the CMCase plate assay [17].
CMCase Assay
A 10 μL aliquot of bacterial broth was inoculated into 4 mm wells made on CMC agar plates. The composition of the CMC agar medium included: Carboxymethylcellulose (CMC), 0.25 g; Tryptone, 2 g; KH2PO4, 4 g; Na2HPO4, 4 g; MgSO4·7H2O, 0.2 g; CaCl2·2H2O, 0.001 g; FeSO4·7H2O, 0.004 g; and Agar, 15 g. The plates were incubated at 37°C for 72 hours. After incubation, the plates were stained with 0.1% Congo red solution for 1 hour, followed by destaining using 1N NaCl solution to detect the cellulose-hydrolyzing zone surrounding the bacterial growth.
Quantitative Assay of Bacterial Cellulase Activity
Cellulase activity was quantified using the DNS method which measures the reducing sugars released by bacterial isolates.
DNS Method
Bacterial isolates exhibiting cellulose-hydrolyzing activity were cultured in nutrient broth and incubated at 37°C overnight. The broths were centrifuged at 10,000 rpm for 10 minutes at 4°C. The substrate solution consisted of 1% CMC dissolved in phosphate buffer (0.1M, pH 6.8). In a 1 ml assay tube, 0.5 ml of the substrate solution and 0.5 ml of the bacterial supernatant (crude enzyme) were combined to form the reaction mixture. The assay tubes were incubated at 55°C for 15 minutes in a water bath. After incubation, 1.5 ml of DNS solution was added to each tube and boiled at 100°C for 10 minutes to stop the reaction. The amount of reducing sugar released was measured spectrophotometrically at 540 nm. A control with 1.5 ml DNS and 1 ml ddH2O was used for calibration [18]. One unit of cellulase activity was defined as the amount of enzyme required to release 1 μmol of glucose per milliliter per minute (Figure 4).
Phenotypic Characterization of Cellulase-Degrading Bacteria
Morphological Characteristics
The morphological and microscopic characteristics of cellulolytic bacteria were examined to determine colony shape, size, spore formation, motility, and Gram’s reaction. Colony characteristics were observed on agar plates, while smears of bacterial cultures were prepared and examined under a phase-contrast microscope using a 100x objective. Gram staining was performed using standard protocols.
Production and Purification of Cellulase Enzyme
Production of Cellulase Enzyme
The most efficient cellulolytic bacterium (BCDB 10) was cultured in a broth medium containing (g/L): K2HPO4, 1.0g; KCl, 0.2g; MgSO4·7H2O, 1.0g; yeast extract, 1.0g. The pH of the culture medium was adjusted to three different values (4.5, 6.75, and 9.0), and various concentrations of Carboxymethylcellulose (CMC) (2.0, 8.5, and 15.0 g/L) were tested to optimize cellulase production. After sterilization, the bacterial strain was inoculated into each flask with a 1% (v/v) inoculum. The flasks were incubated under optimal conditions as determined from previous experiments.
Purification of Cellulase Enzyme
The most potent cellulolytic bacterial strain was cultured in 300 ml of CMC broth, prepared as described earlier. The pH and CMC concentration were adjusted to the optimum values based on Response Surface Methodology (RSM) results [19]. The culture medium was sterilized, and the bacterial strain was inoculated with a 1% (v/v) inoculum. The inoculated flasks were incubated at the optimal temperature and for the optimum incubation period determined from the RSM study, with continuous shaking. After incubation, the supernatant containing crude enzyme was obtained by centrifugation at 10,000 rpm for 10 minutes at 4°C.
The supernatant was then concentrated using a rotary evaporator (Eyela) and subjected to ammonium sulfate precipitation (up to 80%). The enzyme sample was centrifuged at 10,000 rpm for 10 minutes at 4°C to collect the pellet. This pellet was dissolved in a minimal volume of 100 mm phosphate buffer (pH 7.0). The solution was dialyzed overnight using a dialysis membrane with a molecular weight cut-off greater than 5 kDa against the same phosphate buffer.
The dialyzed solution was applied to a DEAE-cellulose column pre-equilibrated with the phosphate buffer. The unbound fractions were collected, and cellulase activity and protein concentration were measured at 540 nm and 280 nm, respectively. Fractions showing the highest cellulase activity were pooled and stored at −20°C for further analysis [20].
In each step of the purification process, the protein concentration was determined using the Lowry method [21], with bovine serum albumin as a standard. To estimate the molecular weight of the cellulase enzyme, SDS-PAGE was performed according to Laemmli’s protocol. The gel consisted of a 12% resolving and 4% stacking gel, and protein bands were visualized using Coomassie Brilliant Blue R-250 staining.
For zymogram analysis, the purified enzyme was subjected to native PAGE with 12% separating and 4% stacking gels. The gel was incubated with 0.5% CMC in phosphate buffer (pH 7.0) for 1 hour, followed by staining with 0.1% Congo red solution for 1 hour. After destaining with 1M NaCl, clear zones indicating cellulase activity were observed around the enzyme bands [22].
Results
Isolation of cellulose degrading bacteria
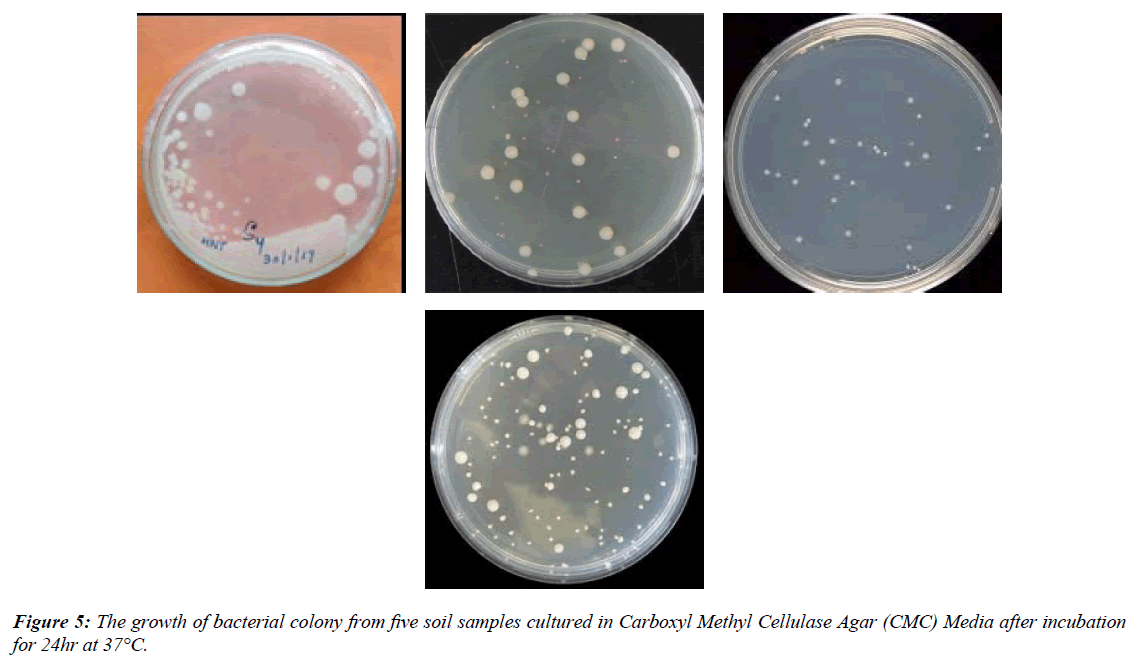
In this study, ten bacterial colonies were isolated from each soil sample using serial dilution and the pour plate technique. The colonies were identified based on morphological characteristics such as shape, size, and color (Figure 5).
Pure culture preparation
For pure culture preparation, morphologically distinct colonies from CMC agar plates were streaked onto nutrient agar plates. After incubation at 37°C for 24 hours, 10 different bacterial isolates grew (Figure 6). A total of 20 pure bacterial isolates were transferred to nutrient agar slants and broth for long-term preservation, named BCDB1 to BCDB20.
Screening of cellulase activity
CMCase plate assay
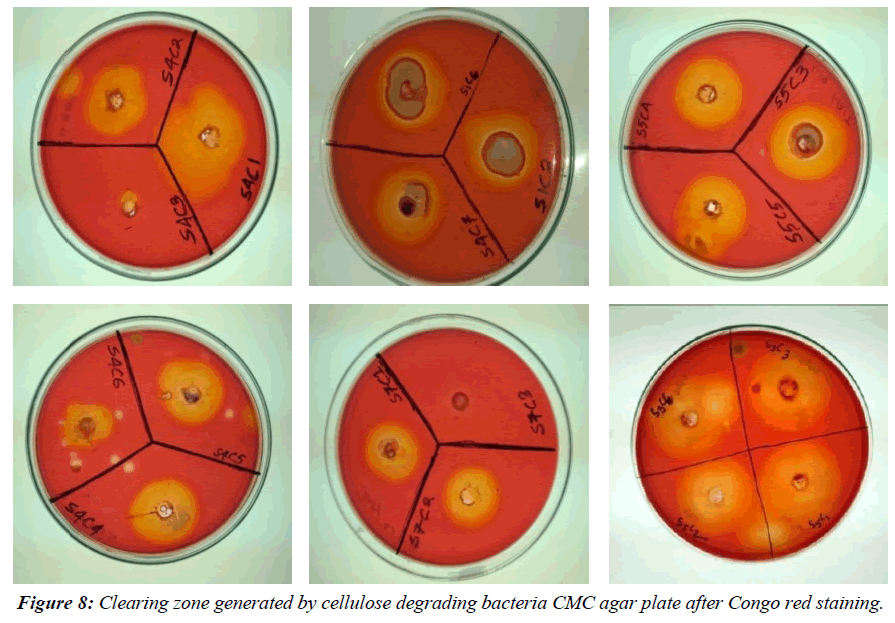
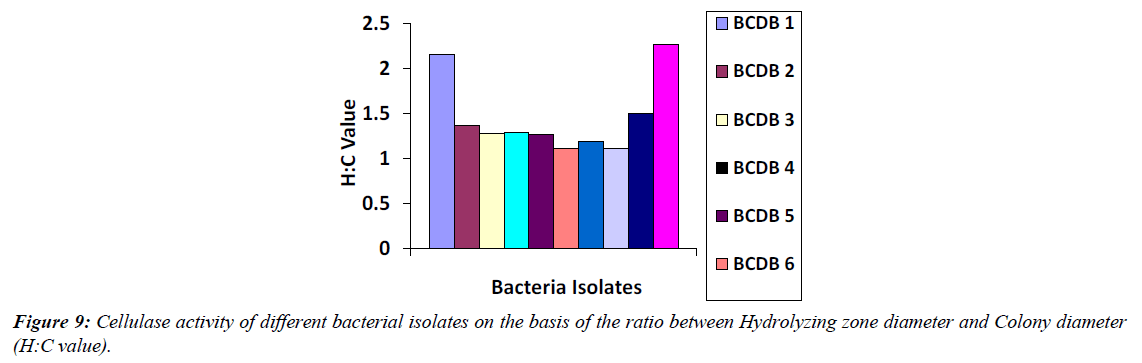
Cellulase activity was assessed using the Carboxymethyl cellulase (CMCase) assay, observing the clear zone formed around bacterial colonies due to enzyme-substrate reactions. Among the 20 bacterial isolates, 10 showed cellulolytic activity with visible hydrolyzing zones on CMC agar plates after staining with 0.1% Congo red and distaining with 1N NaCl (Figure 7-9). Notably, BCDB10, BCDB1, BCDB9, BCDB2, and BCDB3 displayed the most prominent yellow zones, indicating the most efficient cellulase-producing bacterial strains (Table 1).
| Sl No. | Isolate name | Cellulase Activity (halozone) | Colony Diameter* (C) | Hydrolyzing Zone Daiameter* (H) | H:C |
|---|---|---|---|---|---|
| 1 | BCDB 1 | + | 1.3 | 2.8 | 2.153 |
| 2 | BCDB 2 | + | 2.2 | 3 | 1.363 |
| 3 | BCDB 3 | + | 2.5 | 3.2 | 1.28 |
| 4 | BCDB 4 | + | 2.4 | 3.1 | 1.291 |
| 5 | BCDB 5 | + | 2.6 | 3.3 | 1.269 |
| 6 | BCDB 6 | + | 2.7 | 3 | 1.111 |
| 7 | BCDB 7 | + | 2.7 | 3.2 | 1.185 |
| 8 | BCDB 8 | + | 2.8 | 3.1 | 1.107 |
| 9 | BCDB 9 | ++ | 1.8 | 2.7 | 1.5 |
| 10 | BCDB 10 | + | 1.9 | 4.3 | 2.263 |
Table 1: Cellulase activity of different bacterial isolates based on the ratio between Hydrolyzing zone diameter and Colony diameter (H:C Value).
Quantitative assay of bacterial cellulase activity
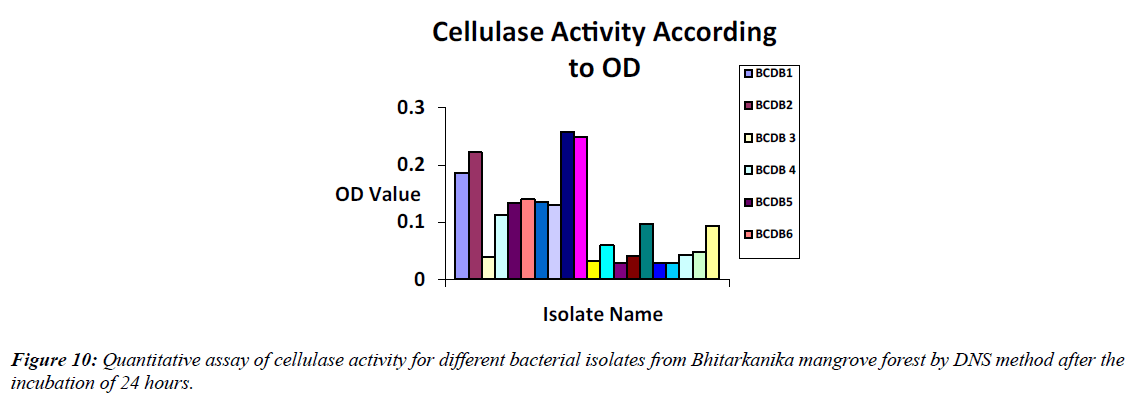
Bacterial isolates BCDB10, BCDB1, and BCDB9 showed the highest cellulase activity in the DNS method, with spectrometric absorbance indicating the maximum reducing sugar released after enzymatic degradation of CMC (Figure 10, Table 2).
| SI No | Isolate name | Cellulase activity according to absorbance |
|---|---|---|
| 1 | BCDB1 | 0.186 |
| 2 | BCDB2 | 0.222 |
| 3 | BCDB 3 | 0.04 |
| 4 | BCDB 4 | 0.113 |
| 5 | BCDB5 | 0.134 |
| 6 | BCDB6 | 0.14 |
| 7 | BCDB 7 | 0.135 |
| 8 | BCDB 8 | 0.13 |
| 9 | BCDB9 | 0.247 |
| 10 | BCDB10 | 0.259 |
| 11 | BCDB 11 | 0.032 |
| 12 | BCDB 12 | 0.059 |
| 13 | BCDB13 | 0.029 |
| 14 | BCDB14 | 0.041 |
| 15 | BCDB 15 | 0.097 |
| 16 | BCDB 16 | 0.028 |
| 17 | BCDB17 | 0.028 |
| 18 | BCDB18 | 0.043 |
| 19 | BCDB 19 | 0.047 |
| 20 | BCDB 20 | 0.093 |
Table 2: Absorbance values of cellulase activity of 20 bacterial isolates from Bhitarkanika mangrove soil by DNS method after the incubation of 24 hours.
Morphological and biochemical characterization of BCDB 10 isolates
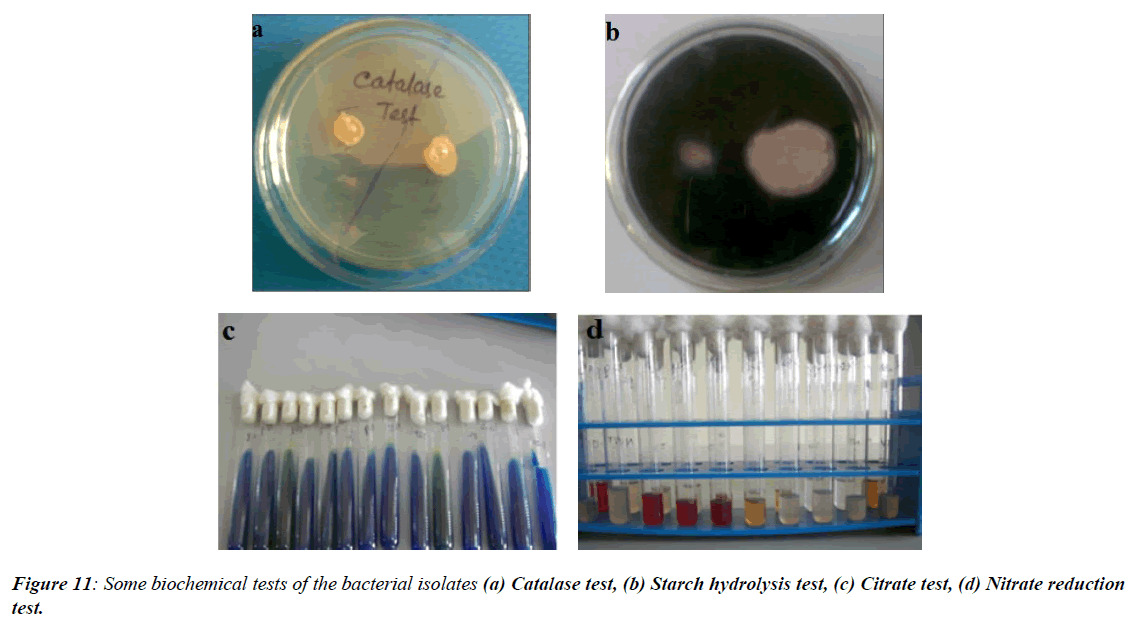
In the present study, bacterial isolate BCDB10 was subjected to morphological characterization, revealing that it was Gram-positive and rod-shaped, as observed under phase contrast microscopy. Several biochemical tests were performed to further identify the cellulolytic bacterium. The results showed that BCDB10 tested positive for urease, methyl red, Voges-Proskauer (VP), and carbohydrate metabolism, while it was negative for oxidase, citrate utilization, catalase, and triple sugar iron tests. Based on these morphological and biochemical characteristics, and by comparing the results with Bergey's Manual of Systematic Bacteriology, BCDB10 was preliminarily identified as a member of the genus Bacillus. These findings suggest that BCDB10 is a Gram-positive, rod-shaped bacterium with notable cellulolytic activity (Figure 11, Table 3).
| Sl .no | Bacterial strain name | Cell morphology | Gram stain |
|---|---|---|---|
| 1 | BCDB 1 | Rod | Positive |
| 2 | BCDB 2 | Rod | Positive |
| 3 | BCDB 3 | Rod | Positive |
| 4 | BCDB 4 | Rod | Positive |
| 5 | BCDB 5 | Cocci | Negative |
| 6 | BCDB 6 | Cocci | Positive |
| 7 | BCDB 7 | Cocci | Positive |
| 8 | BCDB 8 | Rod | Positive |
| 9 | BCDB 9 | Rod | Positive |
| 10 | BCDB 10 | Rod | Positive |
Table 3: Phenotypical characteristics of isolated cellulolytic bacterial strains.
Response surface methodology
In this study, Response Surface Methodology (RSM) was used to optimize cellulase activity of the bacterial strain BCDB10 by examining the effects of environmental factors. Box-Behnken Design (BBD) was employed to establish a second-order polynomial model. The optimized conditions—temperature 37.5°C, pH 6.75, substrate concentration 15 g/L, and incubation time 72 hours—yielded a maximum enzyme activity of 39,250 U/ml. The experimental plan and values are summarized in (Table 4).
| Sl.No. | Incubation (hr) | pH | Substrate conc. (g/L) | Temperature (℃) | Cellulase activity (U/ml) |
|---|---|---|---|---|---|
| 1 | 42 | 9.00 | 15.0 | 37.5 | 47.50 |
| 2 | 72 | 6.75 | 15.0 | 37.5 | 392.50 |
| 3 | 42 | 6.75 | 8.5 | 37.5 | 25.50 |
| 4 | 72 | 6.75 | 2.0 | 37.5 | 10 |
| 5 | 42 | 9.00 | 2.0 | 37.5 | 59.50 |
| 6 | 42 | 6.75 | 2.0 | 30.0 | 88.50 |
| 7 | 72 | 6.75 | 8.5 | 30.0 | 29 |
| 8 | 12 | 6.75 | 2.0 | 37.5 | 52 |
| 9 | 42 | 9.00 | 8.5 | 45.0 | 74.50 |
| 10 | 42 | 6.75 | 15.0 | 45.0 | 300.50 |
| 11 | 42 | 4.50 | 8.5 | 45.0 | 60.50 |
| 12 | 42 | 6.75 | 15.0 | 30.0 | 61.50 |
| 13 | 12 | 6.75 | 15.0 | 37.5 | 56.50 |
| 14 | 42 | 6.75 | 2.0 | 45.0 | 72.50 |
| 15 | 42 | 4.50 | 2.0 | 37.5 | 33 |
| 16 | 12 | 6.75 | 8.5 | 30.0 | 53 |
| 17 | 12 | 4.50 | 8.5 | 37.5 | 80 |
| 18 | 72 | 4.50 | 8.5 | 37.5 | 152.50 |
| 19 | 42 | 6.75 | 8.5 | 37.5 | 19 |
| 20 | 42 | 9.00 | 8.5 | 30.0 | 100.50 |
| 21 | 42 | 4.50 | 15.0 | 37.5 | 13.50 |
| 22 | 72 | 9.00 | 8.5 | 37.5 | 74.00 |
| 23 | 12 | 9.00 | 8.5 | 37.5 | 88.50 |
| 24 | 42 | 4.50 | 8.5 | 30.0 | 79 |
| 25 | 12 | 6.75 | 8.5 | 45.0 | 74 |
| 26 | 42 | 6.75 | 8.5 | 37.5 | 4.50 |
| 27 | 72 | 6.75 | 8.5 | 45.0 | 13.50 |
Table 4: Optimization of cellulase enzyme production.
From the multiple regression analysis, a second-order polynomial equation was derived to predict cellulase production, irrespective of the significance of the coefficients. The response surface model for cellulase activity (Y) is:
Cellulase activity (Y) = 7859.26 + 2229.17*A + 216.67*B + 4637.56*C + 1533.33*D - 2175.00*AB + 9450.00*AC - 912.50*AD + 187.50*BC - 187.50*BD + 6375.00*CD
The coded equation helps predict cellulase activity based on the levels of each factor. It also identifies the relative impact of factors by comparing their coefficients. The model terms A, B, C, D, AB, AC, AD, BC, BD, and CD are significant for cellulase production. The coefficient of variation (CV) value of 99.47 indicates high reliability of the experiment. The p-value of 0.0279 shows that incubation time (A) and substrate concentration (CMC) significantly impact cellulase activity [23].
Purification of cellulase enzyme from Bacillus sp.
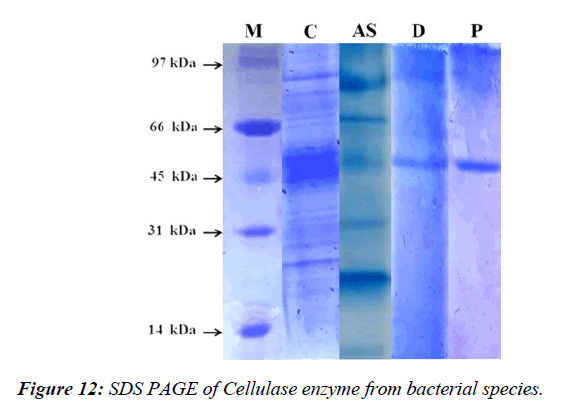
The cellulase enzyme from Bacillus albus was purified using 80% ammonium sulfate precipitation and DEAE-cellulose column chromatography. The enzyme had a molecular mass of approximately 54 kDa, with a 3.98% yield and 5.82-fold purification. A hydrolytic band corresponding to cellulase activity was observed (Figure 12).
Discussion
In the present study, bacterial isolates were obtained from mangrove soil samples collected from Bhitarkanika, Odisha, using CMC-congo red agar medium. The cellulolytic ability of the bacterial isolates was semi-quantitatively assessed by halo zone formation on agar plates. This method revealed the clearing zones around bacterial colonies, indicating cellulose degradation. Fourteen bacterial isolates (CPB 1–14) showed varying degrees of cellulolytic activity. These bacterial isolates exhibited significant cellulose-degrading ability, with their colony growth forming clear halos in the medium containing carboxymethyl cellulose (CMC) as the sole carbon source [24]. The viable plate count indicated that the mangrove soil contained a high number of cellulose-degrading bacteria.
In comparison, He reported the presence of a larger number of cellulose-degrading bacteria in the Sundarbans mangroves in West Bengal, India. The halo zone to colony diameter ratio (HC) values of the isolates ranged from 1.107 (BCDB 8) to 2.263 (BCDB 10). These results were higher than the findings who observed HC ratios between 0.4 and 2.1 for bacterial isolates from wheat farms and forests in Guillan province. Similarly, He observed maximum clearing zones ranging from 2.5 to 6.4 cm, with an average HC value of 4.24 cm, for cellulolytic bacteria isolated from flower stalks and vegetable waste composting systems [25]. However, the HC values from this study were lower than those, who found HC values ranging from 4.85 to 13.11 cm.
Furthermore, the cellulolytic activity of the isolates was also evaluated in broth medium, specifically for their carboxymethyl cellulase (CMCase) activity. The CMCase activity of the 14 isolates was measured after 24 hours of incubation and continued up to 120 hours. The cellulose-degrading efficiency of the isolates ranged from 0.028 (BCDB 16) to 0.259 (BCDB 10). Among these isolates, BCDB 1 and BCDB 10 exhibited the highest cellulase activity and were selected for further study. Notably, BCDB 10 showed maximum cellulase activity after 72 hours of incubation, while BCDB 1 also reached its peak activity at the same time. These findings are consistent with the study which reported cellulase production by various Bacillus species from mangrove soils in the Philippines, ranging from 48.7 to 66.5 U/ml.
Based on their high cellulose-degrading ability, BCDB 1 and BCDB 10 were selected for further identification and optimization. Phenotypic characterization identified the isolates as Bacillus sp. (CPB-3), which was in line with the results, also reported Bacillus cereus, Bacillus licheniformis, Bacillus sp., and Bacillus pumilus from mangrove soils. Similarly [26], He reported the cellulolytic activity of Pseudomonas sp.., Bacillus sp., Bacillus polymyxa, and Bacillus brevis from the mangrove soil of Bhitarkanika, Odisha.
Various environmental parameters such as pH, temperature, and nutrient composition influence the cellulase production by bacteria. In this study, optimization steps were undertaken to identify the optimal conditions for maximal extracellular enzyme production. The results revealed that cellulase production increased gradually up to 72 hours of incubation, after which it declined. This trend aligns with previous studies, who also observed maximum cellulase production by Bacillus sp. at 72 hours.
The pH of the production medium played a crucial role in cellulase production. Both bacterial isolates (BCDB 1 and BCDB 10) showed maximum cellulase production at a pH of 8.0, and enzyme activity decreased significantly when the pH was raised to 9.0. This indicates that these bacterial strains prefer a slightly alkaline pH for optimal cellulolytic activity. This finding aligns with the work of, who reported maximum cellulase production by Bacillus cereus at pH 8.0. They also found that neutral to alkaline pH values are favorable for cellulase production in Bacillus species [27]. In contrast, they reported maximum cellulase production in acidic conditions (pH 6) by Bacillus pumilus isolated from the gut of earthworms.
The optimization of temperature also had a significant impact on enzyme production. Both bacterial isolates produced the highest cellulase activity at 37.5°C, with activity decreasing at higher and lower temperatures. This result is consistent with the findings who observed optimal cellulase production at 37°C for Bacillus subtilis isolated from cow dung. Temperature optimization plays a critical role in enhancing enzyme yield and activity.
In addition to pH and temperature, different carbon and nitrogen sources were tested to assess their effect on enzyme production. The bacterial isolates showed varying preferences for different carbon sources, with CMC being the most effective substrate for cellulase production. Similarly, nitrogen sources like ammonium sulfate and yeast extract enhanced enzyme production. The findings of this study are in line with those who observed the importance of carbon and nitrogen sources in cellulase production by Bacillus subtili [28].
In conclusion, the present study demonstrates that all 10 cellulose-degrading bacterial isolates from the mangrove soil of Bhitarkanika exhibit efficient cellulase production under optimized conditions. These findings are significant for taxonomists, enzymologists, and industrialists working on cellulase production. The use of these cellulose-degrading bacteria as bio-inoculants could enhance organic matter decomposition, improve soil fertility, and reduce the need for chemical fertilizers, promoting sustainable agriculture. Two of the most efficient thermo tolerant cellulose-producing bacterial strains, Bacillus sp. and Pseudomonas sp.., were identified and their cellulase activities were optimized under various growth conditions [29].
The cellulase enzyme from Bacillus sp. was purified using ammonium sulfate precipitation and DEAE-cellulose column chromatography, and it was found to have a molecular mass of approximately 54 kDa. After purification, the enzyme showed a 3.98% yield and a 5.82-fold increase in purity. Further analysis of the purified enzyme revealed optimum activity at pH 7.0 and stability in the pH range of 6.0–8.0, with a relative activity above 60% [30]. These findings suggest the potential application of this enzyme in various industrial processes, such as biofuel production, waste management, and paper recycling.
Conclusion
The present study concludes that all ten cellulose-degrading bacterial isolates from mangrove soil of Bhitarkanika, Odisha, efficiently produce cellulase. Bacillus sp. demonstrated significant hydrolytic efficiency, forming prominent yellow halo zones on CMC-supplemented agar and exhibiting high cellulase activity in the DNSA assay. Characterization of the purified cellulase revealed its potential for industrial applications due to its stability under extreme environmental conditions. However, further detailed studies on enzyme characterization and stabilization are needed to fully explore its industrial potential.
References
- Alvira P, Tomás-Pejó E, Ballesteros M, et al. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: a review. Bio Res Tech. 2010; 101(13):4851-61.
- Balat M, Balat H. Recent trends in global production and utilization of bio-ethanol fuel. Applied Energy. 2009;86(11):2273-82.
- Ballesteros I, Negro MJ, Oliva JM, et al. Ethanol production from steam-explosion pretreated wheat straw. 2006; 496-508.
- Behera M, Dandapat J, Rath CC. Isolation, characterization and screening of bacteria isolates from Similipal Biosphere Reserve forest soil for their metal tolerance capacity and extracellular enzymatic activities. Biorem Biodiv Bioavail. 2009;3:72-8.
- Bhat M. Cellulases and related enzymes in biotechnology. Biotech Adv. 2000;18(5):355-83.
- Binder JB, Raines RT. Simple chemical transformation of lignocellulosic biomass into furans for fuels and chemicals. J Am Chem Soc. 2009;131(5):1979-85.
- Brown Robert C. Biorenewable Resources: Engineering New Products from Agricultural.
- Carrott PJ, Carrott MR, Mourao PA. Pore size control in activated carbons obtained by pyrolysis under different conditions of chemically impregnated cork. J Anal Appl Pyrolysis. 2006;75(2):120-7.
- Carrott PJ, Mourao PA, Carrott MR. Controlling the micropore size of activated carbons for the treatment of fuels and combustion gases. Appl Surf Sci. 2006;252(17):5953-6.
- Champagne P. Bioethanol from agricultural waste residues. Environ Prog. 2008;27(1):51-7.
- Dotaniya ML, Datta SC, Biswas DR, et al. Use of sugarcane industrial by-products for improving sugarcane productivity and soil health. Int J Recycl Org Waste Agricult. 2016;5:185-94.
- Gosselink RJ, De Jong E, Guran B, et al. Co-ordination network for lignin—standardisation, production and applications adapted to market requirements (EUROLIGNIN). Ind Crops Prod. 2004;20(2):121-9.
- Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund MF, et al. Bio-ethanol–the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24(12):549-56.
- Held P. Enzymatic digestion of polysaccharides, part ii: Optimization of polymer digestion and glucose production in microplates. 2012.
- Hemamalini V, Saraswathy SG, Hema C, et al. Comparative study of continuous ethanol fermentation from molasses by using Saccharomyces cervisiae and Schizosaccharomyces pombe. Int J Curr Sci. 2012;1:219-28.
- Heux L, Dinand E, Vignon MR. Structural aspects in ultrathin cellulose microfibrils followed by 13C CP-MAS NMR. Carbohydr Polym. 1999;40(2):115-24.
- Hon DN, editor. Chemical modification of lignocellulosic materials. CRC Press. 1995.
- dos Santos FA, Iulianelli GC, Tavares MI. The use of cellulose nanofillers in obtaining polymer nanocomposites: properties, processing, and applications. Mater Sci Rep. 2016;7(5):257-94.
- Hu J, Arantes V, Saddler JN. The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: is it an additive or synergistic effect?. Biotechnol Biofuels Bioprod. 2011;4:1-4.
- Iwamoto S, Abe K, Yano H. The effect of hemicelluloses on wood pulp nanofibrillation and nanofiber network characteristics. Biomacromolecule. 2008;9(3):1022-6.
- Jena SK, Tayung K, Rath CC, et al. Occurrence of culturable soil fungi in a tropical moist deciduous forest Similipal Biosphere Reserve, Odisha, India. Braz J Microbiol. 2015;46(1):85-96.
- John MJ, Thomas S. Biofibres and biocomposites. Carbohydr Polym. 2008;71(3):343-64.
- Nevell TP, Zeronian SH. Cellulose chemistry and its applications.
- Johnson RK, Zink-Sharp A, Renneckar SH, et al. A new bio-based nanocomposite: fibrillated TEMPO-oxidized celluloses in hydroxypropylcellulose matrix. Cellulose. 2009;16:227-38.
- Khan S, Siddique R, Sajjad W, et al. Biodiesel production from algae to overcome the energy crisis. J Biosci. 2017;24(4):163-7.
- Hort EV, Taylor P. Acetylene‐derived chemicals. 2000.
- Klemm D, Heublein B, Fink HP, et al. Cellulose: fascinating biopolymer and sustainable raw material. 2005;44(22):3358-93.
- Kootstra AM, Mosier NS, Scott EL, et al. Differential effects of mineral and organic acids on the kinetics of arabinose degradation under lignocellulose pretreatment conditions. Biochem. Eng J. 2009;43(1):92-7.
- Krässig H. Cellulose: structure, accessibility, and reactivity Gordon and Breach Sci. 1993.
- Kumar R, Wyman CE. Effect of xylanase supplementation of cellulase on digestion of corn stover solids prepared by leading pretreatment technologies. Bio Res Technology. 2009;100(18):4203-13.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref