Research Article - Biomedical Research (2018) Volume 29, Issue 3
Assessment of the frequency of mitochondrial tRNA variants in patients with ovarian cancer
Ke He1#, Gang Niu1#, Wang Jing1, Lin-Jing Yuan1, Li-Nan Xu2 and Hu Qu3*
1Department of Obstetrics and Gynaecology, the First Affiliated Hospital of Sun Yat-sen University, Guangzhou 510080, PR China
2Department of Reproductive Medicine Research Center, the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou 510655, PR China
3Department of Urology/Reproductive Medicine Research Center, the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou 510655, PR China
#These authors contributed equally
- *Corresponding Author:
- Dr. Hu Qu
Department of Urology/Reproductive Medicine Research
Center The Sixth Affiliated Hospital of Sun Yat-sen University
Guangzhou, PR China
Accepted on November 13, 2017
DOI: 10.4066/biomedicalresearch.29-17-3192
Visit for more related articles at Biomedical ResearchAbstract
Background: Alternations in mitochondrial DNA (mtDNA) have long been proposed to be involved in the pathogenesis of ovarian cancer (OC). However, the allele frequency of mitochondrial tRNA (mttRNA) variants in OC remains largely understood.
Objective: To explore the potential association between mt-tRNA variants and OC.
Methods: 95 OC patients and 50 control subjects were participated in this study. The 22 mt-tRNA genes were PCR amplified and subsequently sequenced by ABI-3700 automatic DNA sequencer. Moreover, we used phylogenetic approach to determine the pathogenic status of these mt-tRNA variants.
Results: We identified 4 mt-tRNA variants: the tRNAThr A15951G; tRNAHis G12192A; tRNAGlu A14693G and tRNALeu(UUR) A3302G, whereas these tRNA variants were absent in healthy controls. Moreover, these variants localized at positions which were highly conserved between different species, and may cause the failure in mt-tRNAs metabolism, consequently result in mitochondrial dysfunction that responsible for OC.
Conclusions: Variations in mt-tRNAs may be associated with OC.
Keywords
Ovarian cancer, Transfer RNA mutations, Mitochondrial dysfunction.
Introduction
Ovarian cancer (OC) is the seventh most common malignancy. It was estimated that in 2012 about 152,000 women died of OC in the world [1]. The incidence of OC in women under the age of 40 years was reported to be 2~14% and had been increasing in recent year [2]. Approximately 70% of patients with OC were diagnosed at an advanced stage, with associated poor prognosis, even after aggressive and immediate treatments. However, up to date, the molecular mechanism underlying OC remained poorly understood [3].
OC cells, similar to other solid tumour cells and in contrast to normal cells, heavily relied on aerobic glycolysis for energy production, a phenomenon known as the Warburg effect [4]. This effect, which was manifested by increased glucose consumption, decreased oxidative phosphorylation (OXPHOS) and accompanying lactate production, had been confirmed and was generally accepted as it had been demonstrated in various tumours. Mitochondria were vital energy-producing organelles in eukaryotic cells that were primarily responsible for generating ATP by OXPHOS. Their efficient functioning was determined by both the nuclear genome and the maternally inherited 16.6-kb mitochondrial genome [5]. Mitochondrial dysfunctions caused by mitochondrial DNA (mtDNA) mutations had been found to be associated with a large number of human cancers, including OC [6]. Owing to the lack of histones protection and a poor DNA repair system, mtDNA had a higher mutation rate than nuclear DNA [7]. Among these, mitochondrial transfer RNA (mt-tRNA) genes were the hot spots for pathogenic mutations associated with OC. However, the frequency of mt-tRNA mutations in OC was not fully elucidated.
In this study, we performed a mutational analysis of mt-tRNA genes in 95 OC patients and 50 healthy subjects using PCR and direct sequence. As a result, we identified 4 potential pathogenic mt-tRNA mutations.
Materials and Methods
Samples
Since January 2015 to December 2016, a total of 95 genetically unrelated Chinese subjects with OC, aged 38 to 66 years old, (average age of 43.6 ± 6.1 years) were recruited in Department of Obstetrics and Gynaecology, the First Affiliated Hospital of Sun Yat-sen University. In addition, 50 control subjects, aged 40 to 65 years old, (average age of 44.1 ± 5.9 years) with ageand body mass index-matched were enrolled in this study. Informed consent, blood samples, and clinical evaluations were obtained from all participants, under protocols approved by the Ethics Committee of the First Affiliated Hospital of Sun Yatsen University. Signed written informed consents were obtained from all the participants before the study. All these patients did not manifestate any other mitochondrial diseases such as deafness, diabetes mellitus, vision loss or neurological disorders.
Screening for the variants in mt-tRNA genes
Genomic DNA was isolated from the whole blood of participants using Puregene DNA Isolation Kits (Gentra Systems, Minneapolis, MN). The fragments spanning all 22 of the mt-tRNA genes of 95 patients and 50 control samples were PCR-amplified by use of sets of the light-strand and the heavy strand oligonucleotide primers as mentioned in a previous investigation [8]. Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using a Big Dye Terminator Cycle sequencing reaction kit. The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_012920) [9].
Structural analysis
The published secondary structures for the mt-tRNAs were used to define the stem and loop structure [10].
Phylogenetic analysis
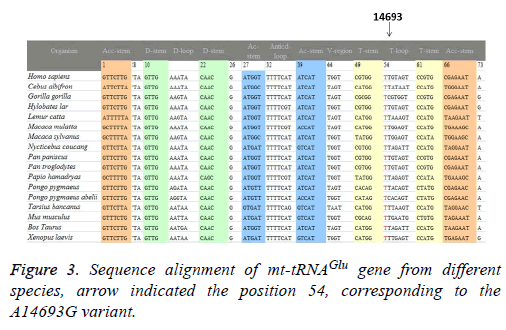
A total of 17 vertebrates’ mtDNA sequences were used in the interspecific analysis. These included Bos taurus, Cebus albifrons, Gorilla gorilla, Homo sapiens, Hylobates lar, Lemur catta, Macaca mulatta, Macaca sylvanus, Mus musculus, Nycticebus coucang, Pan paniscus, Pan troglodytes, Papio hamadryas, Pongo abelii, Pongo pygmaeus, Tarsius bancanus, and Xenopus laevis (Genbank; Table 1). The conservation index (CI) was calculated by comparing the human nucleotide variants with 16 other vertebrates. The CI was then defined as the percentage of species from the list of 17 different vertebrates that had the wild-type nucleotide at that position.
| Species | GenBank Accession Number |
|---|---|
| Homo sapiens | NC_012920 |
| Cebus albifron | NC_002763 |
| Gorilla gorilla | NC_011120 |
| Hylobates lar | NC_002082 |
| Lemur catta | NC_004025 |
| Macaca mulatta | NC_005943 |
| Macaca sylvanus | NC_002764 |
| Nycticebus coucang | NC_002765 |
| Pan paniscus | NC_001644 |
| Pan troglodytes | NC_001643 |
| Papio hamadryas | NC_001992 |
| Pongo pygmaeus | NC_001646 |
| Pongo pygmaeus abelii | NC_002083 |
| Tarsius bancanus | NC_002811 |
| Mus musculus | NC_006914.1 |
| Bos Taurus | HM045018.1 |
| Xenopus laevis | NC_001573.1 |
Table 1. MtDNA sequences of 17 vertebrate species.
Results
Screening for the variants in 22 mt-tRNA genes
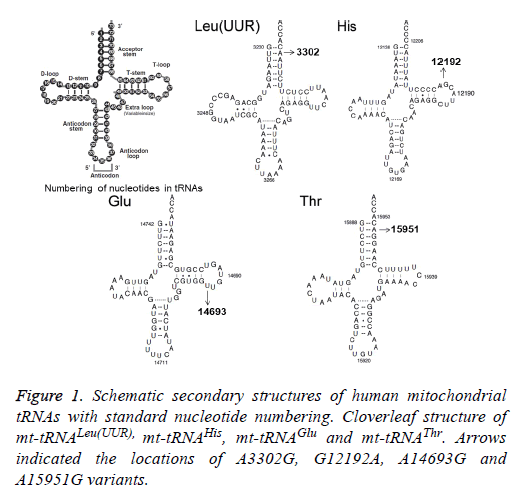
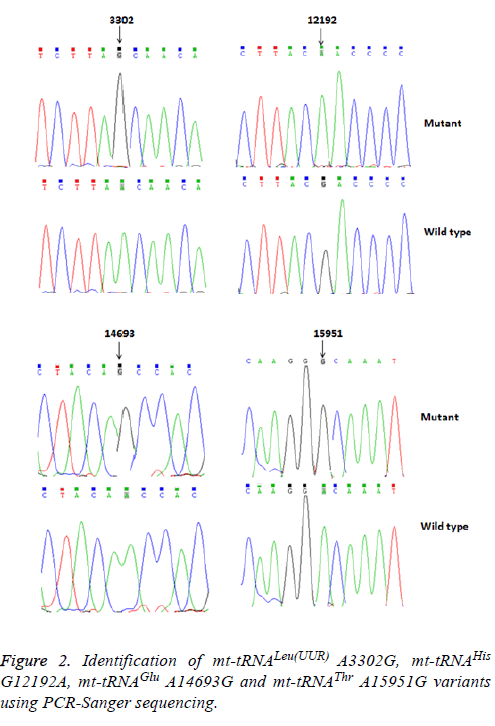
Using PCR and direct sequencing analysis, we identified 6 mttRNA variants: tRNALeu(UUR) A3302G, tRNAHis G12192A; tRNAGlu A14693G; tRNAThr A15951G; tRNAMet T4454C and tRNAArg T10463C (Table 2 and Figure 1).
| Genes | Variant | No. of nucleotide in tRNAs | CI (%) | Homoplasmic/Heteroplasmic | No. of 95 patients (%) |
No. of controls (%) |
|---|---|---|---|---|---|---|
| tRNALeu(UUR) | A3302G | 71 | 100 | Homoplasmic | 1 (1.05) | 0 (0) |
| tRNAHis | G12192A | 59 | 100 | Homoplasmic | 2 (2.1) | 0 (0) |
| tRNAGlu | A14693G | 54 | 100 | Homoplasmic | 1 (1.05) | 0 (0) |
| tRNAThr | A15951G | 71 | 100 | Homoplasmic | 1 (1.05) | 0 (0) |
| tRNAMet | T4454C | 58 | 44.6 | Homoplasmic | 0 (0) | 2 (4) |
| tRNAArg | T10463C | 67 | 31.5 | Homoplasmic | 0 (0) | 1 (2) |
Table 2. Variations in the mt-tRNA genes in 95 subjects with OC and 50 healthy controls.
Of these, the A3302G variant was detected 1 out of 95 OC patients (1.05%); the G12192A variant was detected in 2 patients with OC (2.1%); the A14693G variant was detected in 1 patient (1.05%), the A15951G variant was detected in 1 OC patient (1.05%). In addition, 2 variants were identified only in healthy subjects: the T4454C variant was detected in 2 out of 50 controls (4%); and the T10463C variant was detected in 1 control subject (2%). Notably, the A3302G, G12192A, A14693G and A15951G variants were detected only in the OC patients but were absent in the controls, suggested that these variants may be associated with OC, by contrast, the T4454C and T10463C variants were detected only in controls, suggested that the T4454C and T10463C variants were neutral polymorphisms. The locations of the OC-associated mt-tRNA variants were shown in Figure 2.
Evaluation of OC-associated mt-tRNA pathogenic mutations
To identify putative deleterious mutation, these variants were further evaluated using the following 3 criteria: (1) present in <1% of the controls; (2) CI>75%, proposed by Ruiz-Pesini and Wallace [11]; (3) potential structural and functional alterations. First, we used the secondary structure of mt-tRNAs to localize each variant with either a stem or a loop and to analyze if the base changes within stems altered the classic Watson-Crick base pair. As shown in Figure 2; 2 variants were localized at the acceptor arms, 2 variants occurred at the T arms. Notably, the A3302G and the A15951G variants disrupted the highly conserved Watson-Crick base-pairing. While the G12192A and the A14693G variants created the novel base-pairing in the T arm of the corresponding tRNAs. In addition, a phylogenetic analysis was performed by comparing the human tRNA nucleotide variants with those in 16 other vertebrates. We took the A14693G variant as an example, as shown in Table 2 and Figure 3, the A3302G, G12192A, A14693G and A15951G variants had very high levels of evolutionary conservation (CI=100% for all). Whereas the T4454C and T10463C variants showed lower levels of evolutionary conservation, with the CIs of 44.6% and 31.5%, respectively. Based on these criteria, we proposed that the tRNALeu(UUR) A3302G, tRNAHis G12192A, tRNAGlu A14693G and tRNAThr A15951G variants may have functional significance.
Discussion
In the present study, we screened the potential pathogenic mttRNA mutations in patients with OC. OC was the result of the interaction of multiple genetic and environmental factors, many reports showed that multiples genes may be involved in the pathogenesis of this disease [12-14]. However, these genes were mainly nuclear genes; little was known the role of mitochondrial dysfunction in OC. Since mitochondria used OXPHOS to convert dietary calories into usable energy, releasing reactive oxygen species (ROS) as a toxic by-product. A significant number of epidemiological and clinical studies support the relationship between increased ROS and the pathogenesis of OC [15,16]. Moreover, gene encoding tRNA was the hot spot for pathogenic mutations associated with human mitochondrial diseases. Upto date, over 150 different pathogenic mutations have been reported which are located within mt-tRNA genes [17]. However, the frequency of mttRNA mutations in OC remained largely unknown.
For this purpose, we recently initiated a systematic mutational screening for mt-tRNA genes in 95 patients with OC, as well as 50 control subjects. Sequence analysis of the 22 mt-tRNA genes revealed the presence of 4 variants: tRNALeu(UUR) A3302G, tRNAHis G12192A, tRNAGlu A14693G and tRNAThr A15951G. As shown in Figure 1 and Figure 2, the A3302G variant was localized at the highly conserved acceptor arm of tRNALeu(UUR), disrupted the Watson-Crick base-pairing (2T-71A). It was anticipated that this variant could affect the addition of the CCA triple to the 3’ terminus by the tRNA nucleotidyltransferase. In addition, the A3302G variant was described in patients with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms (MELAS), cardiomyopathy, and myopathy with respiratory insufficiency [18,19]. This variant caused the severe respiratory chain complex I deficiency and lowered complex IV activity. Moreover, a significant reduction in the steady-state level of tRNALeu(UUR) was observed in cell carrying this variant [20]. Studies concerning the functional role of A3302G variant showed that it led to abnormal processing of the mt-16SrRNAtRNALeu( UUR)-ND1 precursor RNA [21]. Therefore, the A3302G should be regarded as a pathogenic mutation associated with OC. However, this mutation was not reported frequently according to the Mitomap database [17].
The G to A transition at position 12192 was first identified in a 60-year-old Japanese man with cardiomyopathy [22]. This variant was located 2 bp from the 3’ end of the T loop of the tRNAHis gene, which was very conserved from bacteria to human mitochondria. In addition, the G12192A had been regarded as a secondary variant for increasing the penetrance and expressivity of Leber’s Hereditary Optic Neuropathy (LHON)-associated ND4 G11778A mutation, however, its allele frequency was very low, since most of the cases were sporadic [23]. In addition, the homoplasmic A14693G variant was localized at the extremely conserved nucleotide of tRNAGlu [24]. Interestingly, the A to G variant at position 14693 occurred at the first base (conventional position 54) of the T loop of tRNAGlu. In fact, nucleotides at position 54 of the T loop of tRNA were often modified, thereby contributing to the structural formation and stabilization of functional tRNAs [25]. Thus, the structural alteration of the tRNA by the A14693G variant may lead to a failure in tRNA metabolism. In a recent study, we noticed that the A14693G variant may modulate the clinical expression of deafness-associated mitochondrial 12S rRNA A1555G mutation [26]. However, its frequency was not very high, we noticed that among 63 deafness pedigree with the A1555G mutation, only 2 subjects carrying this variant [26].
While the A15951G variant was located adjacent to 3’ end, at conventional position 71 of tRNAThr. The adenine (A71) at this position of tRNAThr, highly conserved from bacteria to human mitochondria, had been implicated to be important for tRNA identity and pre-tRNA processing [27]. In addition, ~35% reduction in the level of tRNAThr was observed in cells carrying the A15951G variant. The lower level of tRNAThr in cells carrying the A15951G variant most probably results from a failure to aminoacylate properly this tRNAThr or a defect in the pre-tRNA processing caused by this mutation, as in the case of the A3302G variant in tRNALeu(UUR) [21]. Therefore, the A15951G was a pathogenic variant for OC. However, similar to the A3302G mutation, the allele frequency of the A15951G variant was not frequently.
In summary, our data provided the first evidence for the association between mt-tRNA variants and OC. The main limitation of the current study was the small sample size, further studies including more patients should be performed.
Acknowledgements
This work was supported by the Guangdong Provincial Science and Technology Project (No. 2011B061300048).
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. Parkin DM, Forman D, bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359-386.
- Garg K, Soslow RA. Endometrial carcinoma in women aged 40 years and younger. Arch Pathol Lab Med 2014; 138: 335-342.
- Rauh-Hain JA, Krivak TC, Del Carmen MG, Olawaiye AB. Ovarian cancer screening and early detection in the general population. Rev Obstet Gynecol 2011; 4: 15-21.
- Fabian C, Koetz L, Favaro E, Indraccolo S, Mueller-Klieser W, Sattler UG. Protein profiles in human ovarian cancer cell lines correspond to their metabolic activity and to metabolic profiles of respective tumor xenografts. FEBS J 2012; 279: 882-891.
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet 2005; 6: 389-402.
- Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell 2016; 61: 667-676.
- Picard M, Wallace DC, Burelle Y. The rise of mitochondria in medicine. Mitochondrion 2016; 30:105-116.
- Qin Y, Xue L, Jiang P, Xu M, He Y, Shi S, Huang Y, He J, Mo JQ, Guan MX. Mitochondrial tRNA variants in Chinese subjects with coronary heart disease. J Am Heart Assoc 2014; 3: e000437.
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147.
- Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 2011; 45: 299-329.
- Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum Mutat 2006; 27: 1072-1081.
- Marques DS, Grativol J, Alves da Silva Peres R, da Rocha Matos A, Gimba ERP. Osteopontin-c isoform levels are associated with SR and hnRNP differential expression in ovarian cancer cell lines. Tumour Biol 2017; 39: 1010428317725442.
- Zhu XM, Sun WF. Association between matrix metalloproteinases polymorphisms and ovarian cancer risk: A meta-analysis and systematic review. PLoS One 2017; 12: e0185456.
- Chung VY, Tan TZ, Huang RL, Lai HC, Huang RY. Loss of discoidin domain receptor 1 (DDR1) via CpG methylation during EMT in epithelial ovarian cancer. Gene 2017; 635: 9-15.
- Kim B, Jung JW, Jung J, Han Y, Suh DH, Kim HS, Dhanasekaran DN, Song YS. PGC1α induced by reactive oxygen species contributes to chemoresistance of ovarian cancer cells. Oncotarget 2017; 8: 60299-60311.
- Yin Z, Zhao X, Yang D, Cao X, Yu Y, Jiang H, Zhou X, Li C, Guo Q. LFG-500, a newly synthesized flavonoid, induces apoptosis in human ovarian carcinoma SKOV3 cells with involvement of the reactive oxygen species-mitochondria pathway. Exp Ther Med 2017; 13: 2819-2827.
- Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: A human mitochondrial genome database–2004 update. Nucleic Acids Res 2005; 33: 611-613.
- Goto M, Komaki H, Saito T, Saito Y, Nakagawa E, Sugai K, Sasaki M, Nishino I, Goto Y. MELAS phenotype associated with m.3302A>G mutation in mitochondrial tRNA(Leu(UUR)) gene. Brain Dev 2014; 36: 180-182.
- van den Bosch BJ, de Coo IF, Hendrickx AT, Busch HF, de Jong G, Scholte HR, Smeets HJ. Increased risk for cardiorespiratory failure associated with the A3302G mutation in the mitochondrial DNA encoded tRNALeu(UUR) gene. Neuromuscul Disord 2004; 14: 683-688.
- Maniura-Weber K, Helm M, Engemann K, Eckertz S, Möllers M, Schauen M, Hayrapetyan A, von Kleist-Retzow JC, Lightowlers RN, Bindoff LA, Wiesner RJ. Molecular dysfunction associated with the human mitochondrial 3302A>G mutation in the MTTL1 (mt-tRNALeu(UUR)) gene. Nucleic Acids Res 2006; 34: 6404-6415.
- Bindoff LA, Howell N, Poulton J, McCullough DA, Morten KJ, Lightowlers RN, Turnbull DM, Weber K. Abnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA: a potential disease mechanism. J Biol Chem 1993; 268: 19559-19564.
- Shin WS, Tanaka M, Suzuki J, Hemmi C, Toyo-oka T. A novel homoplasmic mutation in mtDNA with a single evolutionary origin as a risk factor for cardiomyopathy. Am J Hum Genet 2000; 67: 1617-1620.
- Mimaki M, Ikota A, Sato A, Komaki H, Akanuma J, Nonaka I, Goto Y. A double mutation (G11778A and G12192A) in mitochondrial DNA associated with Leber's hereditary optic neuropathy and cardiomyopathy. J Hum Genet 2003; 48: 47-50.
- Florentz C, Sohm B, Tryoen-Tóth P, Pütz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci 2003; 60: 1356-1375.
- Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 2011; 45: 299-329.
- Lu J, Qian Y, Li Z, Yang A, Zhu Y, Li R, Yang L, Tang X, Chen B, Ding Y, Li Y, You J, Zheng J, Tao Z, Zhao F, Wang J, Sun D, Zhao J, Meng Y, Guan MX. Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion 2010; 10: 69-81.
- Li R, Qu J, Zhou X, Tong Y, Hu Y, Qian Y, Lu F, Mo JQ, West CE, Guan MX. The mitochondrial tRNA(Thr) A15951G mutation may influence the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Gene 2006; 376: 79-86.