Research Article - Biomedical Research (2017) Volume 28, Issue 13
An outbreak associated with multidrug-resistant Pseudomonas aeruginosa contamination of duodenoscopes and an automated endoscope reprocessor
Funda Yetkin1*, Yasemin Ersoy1, Cigdem Kuzucu2, Baris Otlu2, Nalan Parmaksiz3 and Yuksel Seckin41Department of Infectious Diseases and Clinical Microbiology, Inonu University School of Medicine, Malatya, Turkey
2Department of Clinical Microbiology, Inonu University School of Medicine, Malatya, Turkey
3Infection Control Nurse, Inonu University School of Medicine, Malatya, Turkey
4Department of Gastroenterology, Inonu University School of Medicine, Malatya, Turkey
- *Corresponding Author:
- Funda Yetkin
Department of Infectious Diseases and Clinical Medicine
Inonu University School of Medicine
Turgut Ozal Medical Center, Turkey
Accepted date: May 31, 2017
Abstract
Background: Duodenoscopes are semi-critical devices used for endoscopic retrograde cholangiopancreatography (ERCP). Disinfection of these instruments is usually based on high-level disinfection procedures with a manual or automated endoscope reprocessor (AER). Duodenoscopes and AER are reported very rarely as a source of infection and outbreaks.
Aim: To investigate an outbreak caused by Pseudomonas aeruginosa in a Gastroenterology Department and ERCP unit in a university hospital and its underlying risk factors.
Method: Three patients in the gastroenterology unit were diagnosed as infected by multidrug-resistant P. aeruginosa and a case control study was conducted for detection of the risk factors. Our infection control team commenced active epidemiological surveillance to determine the cause of these infections. Clonal relationship of the strains was investigated by pulsed field gel electrophoresis (PFGE).
Results: Eight patients were affected in the gastroenterology unit during the period November 2007- February 2008. The case-control analysis confirmed that undergoing ERCP was significantly associated with isolation of P. aeruginosa (P=0.0001) in this unit. Six patients' isolates and seven environmental isolates had an indistinguishable PFGE profile, confirming cross-transmission. The healthcare worker implemented infection control measures to resolve the outbreak and no further cases occurred.
Conclusions: This outbreak resulted from failure of AER and inadequate high level disinfection procedures. AERs can cause contamination of duodenoscopes and can be related P. aeruginosa outbreaks. Reuse of ancillary materials of ERCP play a critical role in outbreak development.
Keywords
Multidrug-resistant P. aeruginosa, Nosocomial infection, Outbreak, Pulsed field gel electrophoresis (PFGE), Endoscopic retrograde cholangiopancreatography (ERCP), Automated endoscope reprocessor (AER).
Introduction
Pseudomonas aeruginosa is a well-known pathogen associated with hospital-acquired infections. Multidrug-resistant P. aeruginosa infections are often associated with increased mortality [1,2]. Specific environmental surfaces identified during outbreak investigations as sources of antibiotic-resistant P. aeruginosa include various medical equipment [3-7]. Endoscopic retrograde cholangiopancreatography (ERCP) is now commonly used in gastroenterology units both for the diagnosis and the treatment of biliary and pancreatic disorders, and outbreaks of ERCP-related infections have been described [8-11]. Although automated endoscope reprocessors (AERs) offer several advantages compared to manual reprocessing, failure of AERs has been linked to outbreaks of infections or colonization [7-9,12-14]. We report an outbreak caused by P. aeruginosa in a university hospital gastroenterology unit and the underlying risk factors.
Materials and Methods
Setting
This study was conducted in a tertiary teaching hospital with 850 beds and 10 intensive care units. The Gastroenterology Department (GD) and ERCP unit are located on the second floor. ERPC is performed on 40 or more patients per month in this unit. Other gastrointestinal endoscopic procedures such as gastroscopy and colonoscopy are performed in the Gastrointestinal Endoscopy Unit on the ground floor. An infection control team, comprising two infectious disease specialists, a microbiologist and three infection control nurses, meets weekly for surveillance and case evaluation.
The gastrointestinal endoscopes and automated endoscope reprocessors
The ERCP unit had three duodenoscopes (two Evis Lucera TJF-260V, one Evis Lucera JF-260V, KeyMed, and Olympus, UK) that are used for retrograde cholangiopancreatography. This duodenoscopes are disinfected in an automated endoscope reprocessor (Choyang CYW-100, Choyang Medical Industry Co., Ltd., Korea) in the ERCP unit. The Gastrointestinal Endoscopy Unit has four gastroscopes (GIF F02-602, Olympus), two colonoscopes (CF 240 L and PCF 240 Olympus), and three AERs (one Choyang CYW-501, Choyang Medical Industry Co., Ltd., Korea, two Choyang CYW-100, Choyang Medical Industry Co., Ltd., Korea). The AERs had been used for 10 months previously. At the end of each procedure, the gastrointestinal endoscopes were cleaned manually with water and detergent before disinfection in the AERs. Each lumen of the endoscopes was scrubbed internally with special brushes. The endoscopes were then disinfected in an AER with 2% glutaraldehyde.
Epidemiological investigation
Case control study: A case-control study was performed to identify the risk factors for a P. aeruginosa outbreak. A “case patient” was defined as any patient who suffered an infection with a P. aeruginosa strain isolated from a clinical specimen between November 2007 and February 2008 in the Gastroenterology Department (GD). “Control patients” were chosen randomly among patients who were hospitalized in this department during the same period but did not develop a P. aeruginosa infection. Each case patient was matched to 7 control patients.
Data collection
The data collected from medical records included age, gender, main diagnosis, underlying diseases, diseases associated with an immune-compromised status, diabetes mellitus, renal failure, heart disease excluding hypertension, previous stay at an intensive care unit (ICU) and antibiotic treatment, microbiological data, clinical evaluation, type of infection, and outcome. Procedure-specific information including date of the procedure and the endoscopes was also recorded. Nosocomial infections were defined according to the criteria of the Centres for Disease Control and Prevention (CDC) [15].
Microbiologic methods and environmental investigations
Environmental sampling: During the outbreak, surveillance samples were taken from various equipment including duodenoscopes, gastroscopes, colonoscopes, biopsy forceps, sphincterotomy knives, and catheters. We also analyzed 19 samples from the surface of the gastrointestinal endoscopy room, the ERCP room, final rinse water from the AERs, the water bottles for endoscopic irrigation, the filters, the detergent tank and the water tank of the AERs and detergent and disinfectant solutions.
Specimens from valves and the washer basin were taken by using pre-moistened sterile cotton swabs. Detergent and disinfectant specimens were taken directly from the detergent tank and unopened detergent bottles. The endoscopes were flushed with 20 ml of sterile water through the biopsy channel and water was collected in sterile containers. Other item sampled included tap water used for manual cleaning and terminal rinsing of endoscopes, and samples from the automatic endoscope reprocessor. All samples were plated onto blood agar and eosin-methylene blue agar.
Identification
P. aeruginosa was identified by its characteristic metabolic test results, and ability to grow at 42ºC. The isolates were further identified by the Phoenix Automated Microbiology System (Becton Dickinson Microbiology System).
Antibiotic susceptibility testing
Minimal inhibitory concentrations (MIC’s) of gentamicin, ceftazidime, ciprofloxacin, levofloxacin, piperacillin/ tazobactam, cefoperazone/sulbactam, cefepime, and imipenem for patients’ isolates were determined by the E Test (bioMerieux, Marcy l'Etoile, France) method. The criteria of the Clinical and Laboratory Standards Institute were used to define susceptibility or resistance to these antimicrobial agents [16].
MDR P. aeruginosa was defined as resistant to at least 3 of the following anti-pseudomonal agents: beta-lactams/inhibitors (piperacillin-tazobactam), cephalosporins (cefepime, ceftazidime), carbapenems (imipenem, meropenem), quinolones (ciprofloxacin, levofloxacin), and aminoglycosides (amikacin, gentamicin) [17].
Genotyping
Pulsed Field Gel Electrophoresis (PFGE): Molecular typing was performed on 15 P. aeruginosa isolates of which 8 were from patient samples and 7 from environmental samples. Isolation and deproteinization of the genomic DNA were done following the rapid PFGE protocol of Durmaz et al. with minor modifications [18]. Briefly, bacterial isolates grown on blood agar were suspended in 1 ml cell suspension buffer (100 mM Tris-HCl, 100 mM EDTA • pH: 8.0•) and the optical density was adjusted to 0.7 (•=590) in the spectrophotometer (Boeco, Hamburg, Germany). The bacterial suspension was mixed with an equal volume of 2% low-melting point agarose (Gibco BRL, Paisley, UK) and dispensed into PFGE plug molds. After enzymatic digestion of the cells and washing of the plugs, genomic DNA in the agarose plugs was restricted by 20 U of SpeI (Promega Corporation, WI, USA). Fragmented DNA was electrophoresed in 1% pulsed-field certified agarose by using a CHEF-DR II system (Bio-Rad Laboratories, Nazareth, Belgium) with the running buffer (0.5 × TBE buffer, pH=8.4) cooled to 14°C at 6 V/cm2 under the following electrophoresis conditions: for block 1, an initial switch time of 5 s, a final switch time of 45 s, and a run time of 20 h; and for block 2, an initial switch time of 30 s, a final switch time of 45 s, and a run time of 10 h. The gel was stained with ethidium bromide (5• g/ml) and photographed under UV light. The DNA band profiles were analyzed with the GelCompar software (version 3.0; Applied Maths, Sint-Martens-Latem, Belgium). The clonal relationships of the strains were evaluated according to the criteria of Tenover et al. [19].
Statistical analysis
SPSS version 16.0 was used for all data management and analysis. The data were expressed as the mean ± SD for continuous variables and as frequencies (%) for categorical variables. The differences of continuous variables between the groups were assessed using the independent sample t test, while the χ2 test was performed for categorical variables. P values<0.05 were considered as statistically significant. Logistic regression analysis was used to examine the relationship between the significant variables. The odds ratios (ORs) were calculated for each explanatory variable.
Results
Description of the outbreak and case patients
During a 24 day period in November 2007, 2 cases of bacteremia and 1 case of peripancreatic abscess caused by multidrug-resistant P. aeruginosa were detected among patients in the GD. The attack rate of P. aeruginosa infections during the outbreak (0.99%) was significantly higher than the attack rate for the former six month period (0.098%; P=0.0078); therefore, an active epidemiological surveillance was commenced. Eight patients were affected during the study period in the GD and our infection control team discovered that all of these cases had recently undergone ERCP. Table 1 shows the demographic and clinical characteristics of the eight definitive case-patients. Four patients developed sepsis either primarily (patients 2 and 6) or secondary to infections at other sites, i.e., cholangitis (patient 3), and empyema of the gallbladder (patient 5). Three of the eight patients (37.5%) (Patients 5, 6 and 7) died. The mortality did not appear to be directly related to the ERCP-associated infection in any of these three patients. Two patients had severe cardiac pathology and one had sepsis syndrome caused by Klebsiella spp.
| Patient No. | Age/sex | Diagnosis | Underlying disease | Date of ERCP | Date isolation | Source of culture | Infection | PFGE type |
|---|---|---|---|---|---|---|---|---|
| 1 | 15/F | Acute pancreatitis | None | 24.10.07 | 07.11.07 | Peripancreatic fluid | Peripancreatic abcess | A |
| 2 | 70/F | Cholangitis | None | 01.11.07 | 06.11.07 | Blood | Primary sepsis | A |
| 3 | 59/M | Choledocholithiasis | None | 07.11.07 | 30.11.07 | Blood | Cholangitis, secondary sepsis | A |
| 4 | 41/M | Cholangitis | None | 12.12.07 | 16.12.07 22.12.07 |
Blood Peritoneal fluid |
Peritonitis | A |
| 5 | 73/F | Choledocholithiasis Cholangitis |
Diabetes mellitus Chronic renal failure Heart failure |
31.12.07 | 03.01.08 | Blood | Empyema of the gallbladder, secondary sepsis | A1 |
| 6 | 85/F | Extrahepatic cholestasis | Coronary artery disease | 27.12.07 | 09.01.08 | Blood | Primary sepsis | A1 |
| 7 | 47/F | Cholecystitis | None | 21.01.08 | 03.02.08 12.02.08 |
Bile Peritoneal fluid |
Necrotizing pancreatitis, secondary sepsis | A |
| 8 | 22/F | Cholecystitis | None | 14.02.08 | 25.02.08 | Peritoneal fluid | Peripancreatic abcess | A |
Table 1: Characteristics of the eight patients infected with multidrug-resistant P. aeruginosa.
Case-control study
The case-control study confirmed that undergoing ERCP recently was significantly associated with isolation of P. aeruginosa in the GD. All 8 case-patients had recently undergone ERCP compared with 14 of the 56 control-patients (100% vs. 25%; P=0.0001), that’s why the OR for this variable was not calculated (Table 2). In addition, a recent stay at the intensive care unit (ICU) and use of antibiotics were also a risk factor for P. aeruginosa infection (OR=16.2; P=0.012 and OR=11.0; P=0.005, respectively).
| Characteristics | Case-Patients (n=8) |
Control-Patients (n=56) |
P | 0R | 95%Cl |
|---|---|---|---|---|---|
| Age (years), mean (SD) | 51.5 (24.8) | 58.0 (16.6) | 0.34 | NA | |
| Female | 5 (62.5%) | 32 (57.1%) | 1.0 | NA | |
| Recently stay in an ICU | 3 (37.5%) | 2 (3.6%) | 0,012 | 16.2 | (2.17-120.92) |
| Prior use of antibiotics | 6 (75.0%) | 12 (21.4%) | 0.005 | 11.0 | (1.96-61.60) |
| Recently ERCP | 8 (100%) | 14 (25%) | 0.0001 | Undefined | |
| Recently esophagogastroscopy | 0 | 21 (37.5%) | 0.045 | NA | |
| Colonoscopy | 0 | 0 | ---- | ---- | |
| Malignancy | 0 | 7 (12.5%) | 0.582 | NA | |
| Hepatobiliary diseases | 8 (100%) | 37 (66.1%) | 0.093 | NA | |
| Diabetes mellitus | 1 (12.5%) | 7(12.5%) | 1.0 | NA | |
| Renal failure | 1 (12.5%) | 3 (5.4%) | 0.422 | NA | |
| Heart disease | 2 (25%) | 7(12.5%) | 0.312 | NA |
Table 2: Characteristics of the cases and controls during the outbreak.
Microbiological investigation of the patients and the environment
P. aeruginosa was isolated from 7 of 35 surveillance samples including three duodenoscopes used for ERCP, rinse water from the AER in the ERCP unit, the water bottles for duodenoscope irrigation and disinfected sphincterotomy knives and catheters.
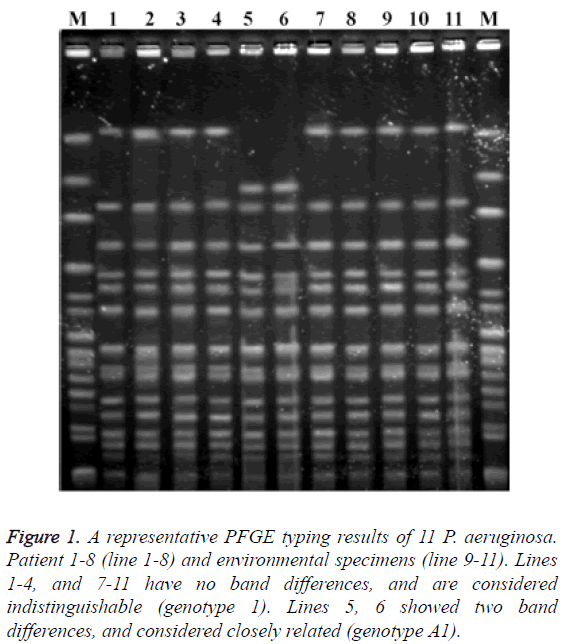
Multidrug-resistance was documented in 85.7% of P. aeruginosa isolated from seven patients. All of these isolates were resistant to imipenem, ceftazidime and the fluoroquinolones. Minimal inhibitory concentration for one isolate could not be determined (isolate lost, not frozen). In addition, the isolates from one patient were susceptible to imipenem, cephalosporins and fluoroquinolones (Table 3). The clonal relationship of strains was shown in Figure 1. The strains of patients 1 and 4 and the strains of patient 7 and one environmental strain had no band differences (genotype 1). Two strains (Patients 5 and 6) had two band differences and considered closely related (genotype A1).
| Minimum Inhibitory Concentration (µg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | N (%) Resistance |
| Gentamicin | 4 | 4 | 4 | 4 | lost | 8 | 4 | 4 | 0 (0) |
| Ceftazidime | 24 | 32 | >32 | 32 | 1 | >32 | 32 | 5 (71.4) | |
| Levofloxacin | >32 | >32 | >32 | 32 | 0.5 | >32 | >32 | 6 (85.7) | |
| Ciprofloxacin | >32 | >32 | >32 | >32 | 12 | >32 | >32 | 7 (100) | |
| Piperacillin/tazobactam | 48 | >256 | 48 | 48 | 4 | 64 | 64 | 1 (14.3) | |
| Sefoperazone/sulbactam | 12 | >256 | 16 | 12 | 4 | 12 | 8 | 1 (14.3) | |
| Sefepime | 12 | >256 | 12 | 8 | 1 | 8 | 12 | 1 (14.3) | |
| Imipenem | >32 | >32 | >32 | >32 | 2 | >32 | >32 | 6 (85.7) | |
Table 3: Antimicrobial susceptibility patterns of the eight P. aeruginosa clinical isolates from the patients.
Figure 1: A representative PFGE typing results of 11 P. aeruginosa. Patient 1-8 (line 1-8) and environmental specimens (line 9-11). Lines 1-4, and 7-11 have no band differences, and are considered indistinguishable (genotype 1). Lines 5, 6 showed two band differences, and considered closely related (genotype A1).
Investigation of the ERCP unit and AERs
When the outbreak broke out, the infection control team visited the ERCP unit and informed ward staff of the problem. All the steps for cleaning and disinfection of the endoscope equipment were checked. The protocol that was prepared previously was not put in practice. Inspection of the duodenoscopes and AER revealed no mechanical damage. The outside of the AER was visually clean. Dark brown debris remained on the scope tip of duodenoscopes. There were no bacteria retaining filter in the AER. Tap water was used to fill the bottle for endoscopic irrigation. Proper cleaning, high-level disinfection and storage of the duodenoscopes were not performed; mechanical scrubbing of the endoscope channel surfaces after the procedure were inadequate, and leak testing was not performed before immersion. After high-level disinfection, the endoscopes were rinsed with tap water and the channels of the endoscopes were not irrigated with alcohol before storage. Finally, the endoscopes were air-dried on a sterile surgical towel and stored horizontally until used again. ERCP accessories (catheters, guide wires, biopsy forceps, and sphincterotomy knives) were cleaned in an ultrasonic cleaner (Olympus Endosonic Ultrasonic Cleaner, Olympus, Japan), packaged without drying, and then immersed in 2% glutaraldehyde for 20 min.
Infection control measures and outcome of the outbreak
Infection control interventions were applied in the unit once an outbreak was suspected. ERCPs were suspended, the contaminated duodenoscopes and AER were withdrawn from service for cleaning and a protocol for endoscope cleaning and disinfection was drawn up for the ERCP unit to overcome the infection. The protocol comprised: (1) cleaning all internal and external surfaces including brushing internal channels and flushing each internal channel with water and an enzymatic detergent, and then rinsing with water (2) immersing the endoscopes in 2% glutaraldehyde, perfusing disinfectant into the channel and exposing it for at least 20 min after leak testing (3) rinsing the endoscope and channels with water (4) flushing the channels of the endoscopes with 70% alcohol and storing by hanging vertically to facilitate air-drying in a storage cabinet. Sterile water was used to fill the bottle for endoscopic irrigation and the water bottle and its connecting tubing underwent high-level disinfection daily. Reusable heat-stable accessories (e.g., biopsy forceps, guide wires) were cleaned with in an ultrasonic cleaner, and then steam sterilized. We recommended that catheters used for ERCP to be “single use”. In addition, all technical staffs were trained regarding cleaning and disinfection procedures for the duodenoscopes and these processes were strictly followed. Bacteriological reassessment was done afterwards, yielding P. aeruginosa in the rinsing water of the AER and these AER devices were cleaned again and remodelled by the manufacturers. The ERCPs were then allowed, and no further case of infection with this strain was detected.
Discussion
Endoscopic retrograde cholangiopancreatography is one of the most invasive endoscopic procedures. ERCP can precipitate infection in the biliary tree and pancreas and carries added risks of peritonitis, cholangitis, and bacteremia. Cholangitis and bacteremia following ERCP can occur due to endogenous bacteria or a contaminated endoscope or accessories [10,20]. Although infectious complications are unusual, contaminated endoscopes used for ERCP have been the sources of nosocomial transmission of bacterial agents. We have described an outbreak of MDR-resistant P. aeruginosa among patients who underwent ERCP in the GD. We considered cross-contamination related to inadequate disinfection of duodenoscopes and accessories used for ERCP. P. aeruginosa was isolated from seven surveillance samples including all duodenoscopes used for ERCP, rinse water from the AER in the ERCP unit, the water bottles for duodenoscope irrigation and disinfected sphincterotomy knives and catheters. Genotypic analysis by PFGE revealed that 6 patients’ isolates and 7 environmental isolates were of the same pattern (Genotype A) and 2 patients’ isolates were found to be clonally related to those patterns (Genotype A1). The case–control study confirmed that recent exposure to ERCP was associated with a P. aeruginosa infection (P=0.0001, OR: Undefined). In agreement with previous data, our study also showed that recent stay at the ICU and prior uses of antibiotics were significantly associated with the isolation of P. aeruginosa [21].
There have been several reports of P. aeruginosa outbreaks attributed to inadequate cleaning and disinfection procedures of instruments used for ERCP [8-11,20]. These outbreaks were related to a number of problems, including inadequate cleaning or disinfection of endoscope channels and accessories, insufficient drying of channels, or using flawed AER.
P. aeruginosa is one of the main organisms responsible for drug-resistant nosocomial infections. Outbreaks of MDR P. aeruginosa in hospitals are an increasing infection control problem [4,11]. Antibiotic resistance has also increased in our hospital and country [22,23]. The incidence of infections due to these resistant strains is associated with increased morbidity, mortality, and cost [17,24]. In our study, multidrug-resistance was documented in 85.7% of P. aeruginosa isolates and this condition caused serious problems in the therapeutic management of patients.
The complexity of endoscopes makes them difficult to clean. These difficulties have been exploited by organisms such as P. aeruginosa, a pathogen ideally suited to take advantage of an inadequately disinfected endoscope [11]. There is, however, a downside to the use of automated washer/disinfectors, which relates to recontamination of endoscopes by the machine after the disinfection. This is due to the growth of microorganisms within a biofilm present in the tanks and pipes of the washer/ disinfector and thus recontamination during the final rinse prior to removal of the instrument from the machine [25]. In addition some endoscopes such as the duodenoscopes used for ERCP contain features (e.g., elevator-wire channel) that require a flushing pressure that is not achieved by most AERs [26]. Initial manual cleaning is therefore vital to the whole decontamination process and is still necessary before the endoscope reprocessor can be used; it must not be neglected [12].
The use of AERs offers several advantages over manual cleaning. The process standardizes several important reprocessing steps and also reduces worker contact with contaminated items and personnel exposure to high-level disinfectants or chemical sterilisers [12,13]. Failure of AERs has been linked to outbreaks of healthcare-associated infections [7-9,14]. Allen et al. described an outbreak of serious P. aeruginosa infections related to inadequate decontamination of the ERCP endoscope, recontamination from the rinse water of the AER, and inadequate drying of the endoscope by the AER [8]. Struelens et al. showed that the post-ERCP infection rate increased from 1.6% to 3.6% following the use of a new automated washer/disinfector while effective decontamination of the washer/disinfector led to a reduction in the infection rate to pre-incident levels [20]. A recent report identified contamination by P. aeruginosa in the detergent tank of the AER [7]. We also identified contamination by P. aeruginosa in the AER. This probably was due to the growth of microorganism within a biofilm present in the pipes of the AER. Bacteriologic reassessment was done afterwards, yielding P. aeruginosa in the rinsing water of the AER and these AERs were cleaned again and remodelled by the manufacturers.
Outbreaks involving removable endoscope parts such as suction valves and endoscopic accessories have been described. Because of the complexity of reprocessing, many of these devices have been designed for single use. Particularly those used for ERCP are considered critical as they enter normally sterile body cavities [27]. However, reprocessing of ERCP accessories is a vital issue in developing countries. Reuse of single-use accessories after adequate reprocessing is a controversial issue. The reprocessing of endoscopic accessories remains an unresolved issue, essentially because the cost of these accessories is considerable, and economic forces dictate the need to reuse many of them [12,27]. Thus, it has been suggested that small centres that do not have a high endoscopy load might reuse accessories after adequate cleaning followed by disinfection in glutaraldehyde for 10 min [12].
We controlled the outbreak with an intervention of sterilization of accessories or single use of catheter or other accessories of the duodenoscope and applying the disinfection protocols for the duodenoscope. However, the AER cultures were sometimes positive in the period after the outbreak. We stopped using an AER for the disinfection of duodenoscopes. There was no case linked to endoscopy procedures afterwards.
In conclusion, AERs can cause contamination of duodenoscopes and can be related to a P. aeruginosa outbreak. There is a need for further redesign of AERs and development of duodenoscopes so that they do not lead to infections. In addition, the reprocessing of endoscopic accessories remains a contentious issue particularly in developing countries, as the cost of these accessories is considerable. Ancillary materials of ERCP should be used sterile if possible as reuse can cause serious problems.
Acknowledgments
All authors express their sincere gratitude to Saim Yologlu, from the Department of Biostatistics, School of Medicine, Inonu University for his help in statistical analysis of this manuscript.
Potential Conflicts of Interest
All authors report no conflicts of interest relevant to this article.
References
- Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Kim EC. Risk factors for antimicrobial resistance and influence of resistance on mortality in patients with bloodstream infection caused by Pseudomonas aeruginosa. Microb Drug Resist 2005; 11: 68-74.
- Linden PK, Kusne S, Coley K, Fontes P, Kramer DJ, Paterson D. Use of parenteral colistin for the treatment of serious infection due to antimicrobial-resistant Pseudomonas aeruginosa. Clin Infect Dis 2003; 37: e154-e160.
- Berrouane YF, McNutt LA, Buschelman BJ, Rhomberg PR, Sanford MD, Hollis RJ. Outbreak of severe Pseudomonas aeruginosa infections caused by a contaminated drain in a whirlpool bathtub. Clin Infect Dis 2000; 31: 1331-1337.
- Bukholm G, Tannaes T, Kjelsberg AB, Smith-Erichsen N. An outbreak of multidrug-resistant Pseudomonas aeruginosa associated with increased risk of patient death in an intensive care unit. Infect Control Hosp Epidemiol 2002; 23: 441-446.
- Kikuchi T, Nagashima G, Taguchi K, Kuraishi H, Nemoto H, Yamanaka M. Contaminated oral intubation equipment associated with an outbreak of carbapenem-resistant Pseudomonas in an intensive care unit. J Hosp Infect 2007; 65: 54-57.
- Pena C, Dominguez MA, Pujol M, Verdaguer R, Gudiol F, Ariza J. An outbreak of carbapenem-resistant Pseudomonas aeruginosa in a urology ward. Clin Microbiol Infect 2003; 9: 938-943.
- Shimono N, Takuma T, Tsuchimochi N, Shiose A, Murata M, Kanamoto Y. An outbreak of Pseudomonas aeruginosa infections following thoracic surgeries occurring via the contamination of bronchoscopes and an automatic endoscope reprocessor. J Infect Chemother 2008; 14: 418-423.
- Allen JI, Allen MO, Olson MM, Gerding DN, Shanholtzer CJ, Meier PB. Pseudomonas infection of the biliary system resulting from use of a contaminated endoscope. Gastroenterol 1987; 92: 759-763.
- Alvarado CJ, Stolz SM, Maki DG. Nosocomial infections from contaminated endoscopes: a flawed automated endoscope washer. An investigation using molecular epidemiology. Am J Med 1991; 91: 272S-280S.
- Classen DC, Jacobson JA, Burke JP, Jacobson JT, Evans RS. Serious Pseudomonas infections associated with endoscopic retrograde cholangiopancreatography. Am J Med 1988; 84: 590-596.
- Fraser TG, Reiner S, Malczynski M, Yarnold PR, Warren J, Noskin GA. Multidrug-resistant Pseudomonas aeruginosa cholangitis after endoscopic retrograde cholangiopancreatography: failure of routine endoscope cultures to prevent an outbreak. Infect Control Hosp Epidemiol 2004; 25: 856-859.
- Ramakrishna BS. Safety of technology: infection control standards in endoscopy. J Gastroenterol Hepatol 2002; 17: 361-368.
- Rutala WA, Weber DJ, HICPAC. Guideline for Disinfection and Sterilization in Healthcare Facilities. CDC Guideline 2008; 1-158.
- Schelenz S, French G. An outbreak of multidrug-resistant Pseudomonas aeruginosa infection associated with contamination of bronchoscopes and an endoscope washer-disinfector. J Hosp Infect 2000; 46: 23-30.
- Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control 1988; 16: 128-140.
- Clinical and Laboratory Standard Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI document M100-S19CLSI (edn). Wayne, PA, USA.
- Tumbarello M, Repetto E, Trecarichi EM, Bernardini C, DE PG, Parisini A. Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: risk factors and mortality. Epidemiol Infect 2011; 1-10.
- Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, Ersoy Y. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis 2009; 62: 372-377.
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995; 33: 2233-2239.
- Struelens MJ, Rost F, Deplano A, Maas A, Schwam V, Serruys E. Pseudomonas aeruginosa and Enterobacteriaceae bacteremia after biliary endoscopy: an outbreak investigation using DNA macrorestriction analysis. Am J Med 1993; 95: 489-498.
- Falagas ME, Kopterides P. Risk factors for the isolation of multi-drug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: a systematic review of the literature. J Hosp Infect 2006; 64: 7-15.
- Gonlugur U, Bakici MZ, Ozdemir L, Akkurt I, Icagasioglu S, Gultekin F. Retrospective analysis of antibiotic susceptibility patterns of respiratory isolates of Pseudomonas aeruginosa in a Turkish University Hospital. Ann Clin Microbiol Antimicrob 2003; 2: 5.
- Yetkin G, Otlu B, Cicek A, Kuzucu C, Durmaz R. Clinical, microbiologic, and epidemiologic characteristics of Pseudomonas aeruginosa infections in a University Hospital, Malatya, Turkey. Am J Infect Control 2006; 34: 188-192.
- Giske CG, Monnet DL, Cars O, Carmeli Y. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob Agents Chemother 2008; 52: 813-821.
- Maloney S, Welbel S, Daves B, Adams K, Becker S, Bland L. Mycobacterium abscessus pseudoinfection traced to an automated endoscope washer: utility of epidemiologic and laboratory investigation. J Infect Dis 1994; 169: 1166-1169.
- Muscarella LF. Automatic flexible endoscope reprocessors. Gastrointest Endosc Clin N Am 2000; 10: 245-257.
- Ogoshi K. Reprocessing of gastrointestinal endoscopic accessories. J Gastroenterol Hepatol 2000; 15 Suppl: G82-G85.