Research Article - Biomedical Research (2018) Volume 29, Issue 13
A study of CYP2C19*2, *3 and *7 in different Sudanese ethnic groups and their response to omeprazole based triple therapy in Khartoum, Sudan 2016-2017
Safinaz Ibrahim Khalil* and Abdalla Omer Elkhawad
Department of Clinical Pharmacology, University of Medical Sciences and Technology, Khartoum, Sudan
- *Corresponding Author:
- Safinaz Ibrahim Khalil
Department of Clinical Pharmacology
University of Medical Sciences and Technology
Khartoum, Sudan
Accepted date: June 06, 2018
DOI: 10.4066/biomedicalresearch.29-18-727
Visit for more related articles at Biomedical ResearchAbstract
Background: Peptic ulcer disease has a high burden worldwide and the commonest causative agent is H. pylori which are usually eradicated with omeprazole-based triple therapy. Pharmacogenetics is an important branch of pharmacology and should be applied to assist the clinical usage of medicines which has a strong relation with enzyme activity of certain genes and alleles known through the literature. CYP2C19 is known to affect the activity of omeprazole and this will result in different responses to treatment. The commonest alleles through the literature are CYP2C19*2, and CYP2C19*3 which vary according to different ethnicity of different populations.
Aim: In Sudan, there are versatile ethnic groups which might affect the response to treatment by many drugs dependent genotypes for example CYP2C19 and Omeprazole interaction and hence studies in this area should be done as they are few.
Methodology: A purposeful convenient sampling; in which patients with peptic ulcer disease and treated with omeprazole were reviewed and the PCR is used to differentiate the CYP2C19 different alleles in patients presenting from February 2016 to January 2017.
Results: In the present study we investigated the distribution of three common gene variants affecting the omeprazole treatment of peptic ulcer disease and H. pylori eradication namely to CYP2C19*2, *3 and *7. The CYP2C19*2 mutation was found among all seven ethnic groups of Sudan, Arabs mostly 17 (139) followed by Darforians 9 (139), Beja 6 (139) and Nilotics 6 (139), Nuba 5 (139) then, Nubians 3 (139) and Fulani 1 (139), p=0.048 which is significant. Regarding CYP2C19*3 mutation of this allele is found in certain ethnic groups Arabs 6 (139), Nubians 6 (139) and Nuba 2 (139), p=0.043 significant. There is no mutation found among different Sudanese ethnic groups in CYP2C19*7. Arabs are normal homozygotes 6 (139) and Darforians 1 (139). Nuba 1 (139) were found to be heterozygotes in this allele p=0.038 significant. Treatment of H. pylori with omeprazole-based triple therapy was used in 110 (139) and no significant correlation found with the different ethnic groups of Sudan.
Conclusion: These data indicate that Sudanese seven ethnic groups showed activity of CYP2C19*2, CYP2C19*3 were they took omeprazole based triple treatment and some of them showed activity to CYP2C19*7.
Keywords
Peptic ulcer disease, Omeprazole, Sudan ethic groups, CYP2C19*2, *3, *7
List of Abbreviations
ATPase: Adenosine Triphosphate; AP-1: Activator Protein-1; Cag A: Cytotoxin associated gene A; CYP2C19: Cytochrome P450 2C 19; DNA: Deoxy Ribonucleic Acid; ELISA: Enzyme- Linked Immunosorbent Assay; FDA: Food and Drug Administration; H. pylori: Helicobacter pylori; NSAIDS: Non- Steroidal Anti-Inflammatory Drugs; PAI: Pathogenicity Island; PM: Poor Metabolizer; IM: Intermediate Metabolizer; RM: Rapid Metabolizer; UM: Ultra-rapid Metabolizer; PPI: Protein Pump Inhibitor; UBT: Urea Breath Test; WHO: World Health Organization; AUC: Area Under the Curve.
Introduction
Peptic Ulcer Disease (PUD) consists of either gastric or duodenal type. In both, the patient usually presents with epigastric pain. Other manifestations are dyspepsia, heartburn, hematemesis or melena, briskly bleeding ulcer, anaemia and perforated ulcer.
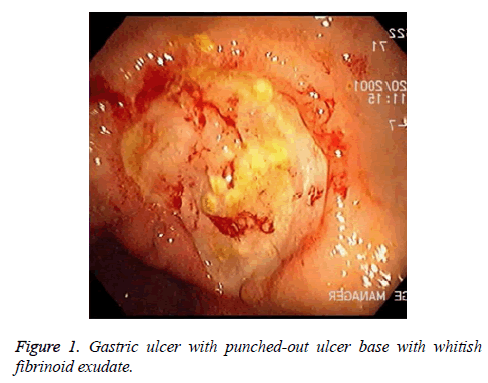
There are alarm symptoms that warrant referral to the gastroenterologist, such as bleeding, anaemia, early satiety, unexplained weight loss, progressive dysphagia or odynophagia, recurrent vomiting and family history of GI cancer. The most helpful test for diagnosing peptic ulcer disease is by radiographic or endoscopic confirmation. H. pylori testing are a must for all patients. The best tool for the diagnosis is the endoscopic rapid urease test. Evaluation of patients suspected of the disease. Doctors usually prefer to do upper GI endoscopy (Figure 1).
Treatment of choice for the peptic ulcer disease with cure of H. pylori infection in about 80-95%, with PPI-based triple therapy; there is a high recurrence rate if H. pylori infection is not successfully treated. Risk factors which are known to affect the PUD are cigarette smoking, advanced age, former alcohol use, obesity and specific chronic diseases.
Omeprazole and CYP2C19
Cytochrome P450 (CYP) enzymes, present in the liver in great amount, regulate the metabolism of several prescription drugs including proton pump inhibitor. CYP2C19 is an isoenzyme which mediates the metabolic activation and elimination, and therefore the therapeutic effect of many drugs including omeprazole.
Cytochrome P450 (CYP) enzymes are mainly found in the liver, their main function is to regulate the metabolism of several prescription drugs including Omeprazole.CYP2C19 is an isoenzyme that mediates the metabolic activation and elimination, and hence the therapeutic effect of many drugs including omeprazole. The ability to metabolize drugs that are CYP2C19 substrates differs among patients resulting in wide variability in their response to specific drug therapy [1].
The omeprazole is a proton pump inhibitor it acts through blocking the gastric acid secretion. It is used for treating Gastroesophageal Reflux Disease (GERD), gastric ulcer, duodenal ulcer, erosive gastritis and other acid-related disorders. It is used also in treating hyper-secretory conditions, such as Zollinger Ellison syndrome, and is used in combination with antibiotics to eradicate Helicobacter pylori infection [1].
PPIs are commonly prescribed for certain conditions like gastroesophageal reflux disease, gastric and duodenal ulcer disease, eradication of Helicobacter pylori (H. pylori) infection, prevention and treatment of non-steroidal antiinflammatory drug-associated damage, and for patients with non-variceal upper gastrointestinal bleeding or non-nuclear dyspepsia. Most PPIs are predominantly metabolized by CYP2C19, both IMs and PMs can have reduced drug elimination and higher PPI plasma concentrations compared with EM individuals [2]. Therefore, eradication of H. pylori infection with omeprazole, lansoprazole, and pantoprazole has been reported to be greater among CYP2C19 IMs and PMs compared with EMs [2,3]. Additionally, the healing rates of peptic ulcers and gastroesophageal reflux disease during PPI treatment is influenced by CYP2C19 genotype as IMs and PMs have been found to respond better to PPI treatment than EMs [3-6].
The UM genotype (i.e., *17/*17) has been reported to affect omeprazole pharmacokinetics resulting in increased rates of drug metabolism and sub-therapeutic exposure [7]. Although, not all studies have identified a significant effect of CYP2C19*17 on PPI metabolism and H. pylori eradication [8,9].
Cause of such inter-individual variability is associated with variations in CYP2C19gene encoding CYP2C19 enzyme. Presence of a specific genetic variant depends on racial and ethnic background [10]. Around 25 variants have been identified for the CYP2C19 gene, and about a dozen variants, which are functionally impaired, can lead to the production of CYP2C19 with high, low or no enzyme activity causing abnormal levels of drugs and their derivatives. This will result in adverse events which include toxicity, sub-therapeutic effect or non-responsiveness. The CYP2C19 isoenzymes metabolizes approximately 5-10% of all prescription drugs, and poor metabolizers taking those drugs may be predisposed to side effects due to under-dosing or over-dosing, depending on whether it is a pro-drug or active drug respectively [11].
The genetic test is used to identify patients at risk for adverse effects due to impaired metabolism by: Detection 6 variants of CYP2C19 gene responsible for non-functional metabolic activity CYPC2C19*2, *3, *4, *6, *7, and *8. Detection 2 variants of CYP2C19 gene responsible for a decrease in functional metabolic activity CYPC2C19*9 and *10; detection one variant of CYP2C19 gene responsible for increased functional metabolic activity CYP2C19 *17 [12].
A *2 or *3 result for CYP2C19 associated to a poor metabolizer phenotype for all drugs metabolized by CYP2C19. For CYP2C19*4, *5, *6, *7 alleles are found to impair metabolism of omeprazole although they are less frequent [13].
Non-genetic factors such as concurrent medications, impaired hepatic function, obesity, insulin resistance and noncompliance can also affect CYP2C19 metabolism. These can lead to decrease or increase in CYP2C19 function in relation to the suggested genotype. Co-administration of drugs metabolized by CYP2C19 may increase or decrease the CYP219 activity. Common drugs involved in the metabolism by CYP2C19 and decrease tier activity and though their toxicity are chloramphenicol, cimetidine, delavirdine, efavirenz, felbamate, fluconazole, fluoxetine fluvoxamine, indomethacin, isoniazid, ketoconazole, lansoprazole, modafinil, omeprazole, oral contraceptives, oxcarbazepine, probenecid, ticlopidine, topiramate, voriconazole (Table 1) [12,14-17].
| Genotype | Allele | Phenotype |
|---|---|---|
| CYP2C19*1 | wild type | EM/Normal function |
| CYP2C19*2 | 681 G>A | PM/Non-function |
| CYP2C19*3 | 636 G>A | PM/Non-function |
| CYP2C19*4 | 1 A>G | PM/Non-function |
| CYP2C19*6 | 395 G>A | PM/Non-function |
| CYP2C19*7 | IVS 5+2T>A | PM/Non-function |
| CYP2C19*8 | 358 T>C | PM/Non-function |
| CYP2C19*9 | 431 G>A | IM/Decreased function |
| CYP2C19*10 | 680 C>T | IM/Decreased function |
| CYP2C19*17 | -806 C>T | UM/Increased function |
Table 1. Cytochrome P450 2C19 genotype and phenotype for drug metabolism.
The ability to metabolize drugs that are CYP2C19 substrates differs among patients resulting in versatile variability in their response to specific drug therapy. A responsible cause of such inter-individual variability is associated with variations in CYP2C19 gene encoding CYP2C19 enzyme. Prevalence of a specific genetic variant depends on racial and ethnic background. More than 25 variants have been identified for the CYP2C19 gene, and about a dozen variants, which are functionally impaired, can lead to the production of CYP2C19 with high, low or no enzyme activity causing abnormal levels of drugs. This can result in adverse events that include toxicity, sub-therapeutic effect or non-responsiveness [1]. Prevalence of CYP2C19 gene variants differs depending on ethnic background. The frequency of allele CYP2C19*2 has been reported as approximately 30-35% of Asians, and 15-26% of Caucasian and African Americans. The frequency of allele CYP2C19*3 has been reported as approximately 10% of Asians, and less than 2% of Caucasian and African Americans. African individuals are the most ancient population therefore they have exposed to greater genomic diversification than non- African populations [18-21].
Sudan is one of the largest African countries with many different ethnic groups and is difficult to race out right Sudan due to the intermarriage between various indigenous and immigrant’s groups. The ethnicity can be categorized in 7 main groups: 39% claims an Arab descent, 30% are of African origin, 12% are Beja, 15% are Nubian and 4% other [1].
Justification and hypothesis
Regarding that omeprazole based triple therapy is not effective for eradication of H. pylori in many hospitals in Sudan and the recurrence of the peptic ulcer disease in these patients is very common as claimed by many gastroenterologists. Is there a relation between the CYP2C19 activity in Sudanese different ethnic groups a relation with the therapeutic failure rate?
Failure of treatment by reviewing the literature, omeprazole is one of the approved drugs to be affected by CYP2C19*2 and *3 [2,19].
Hypothesis
Accordingly, due to the lack of knowledge on the specific gene of interest (CYP2C19) we would like to investigate the differences in omeprazole action against peptic ulcer disease among selected Sudanese ethnic groups. The importance of conducting such a study is stemming out of the fact that high percentage of the African including Sudan are suffering from the disease.
To study CYP2C19*2, *3 and *7 activity among different Sudanese ethnic groups who have peptic ulcer disease and treated with omeprazole based triple treatment.
Objectives
General objectives
To study of CYP2C19*2, *3 and *7 in different Sudanese ethnic groups and their response to Omeprazole based triple therapy using molecular technique.
The specific objectives of the study
1. To study the urease positivity to H. pylori gene among the selected study group by polymerase chain reaction (PCR).
2. To study the frequency of CYP2C19*2, CYP2C19*3 and CYP2C19*7 among different Sudanese ethnic groups.
3. To study the relation between the different ethnic groups of Sudan and CYP2C19*2, CYP2C19*3 and CYP2C19*7 normal homozygotes, mutant, and heterozygotes.
4. To study the response of different ethnic groups of Sudan to omeprazole based triple treatment.
Literature Review
Pathophysiology
In peptic ulcer disease there is a defect in the gastric and duodenal mucosa covering all the muscularis mucosa. Stomach epithelial cells and duodenum secrete mucus in response to irritation of the epithelial lining and cholinergic stimulation as well.
In the epithelial cells, there is the basolateral cell membrane ion pumps aid in the regulation of the intracellular pH where it removes excess hydrogen ions.
Normal physiological balance exists between gastric acid secretion and gastroduodenal mucosal defence. When this balance is disturbed then peptic ulcer disease will occur. NSAIDs, H. pylori infection, alcohol, bile salts, acid and pepsin are aggressive factors that disturb this physiological balance.
The primary causative agent of peptic ulcer disease is the H. pylori. It is a gram-negative spirochete, linked to gastritis since 1983 [22]. What makes it survive for a long time is that it alkalinizes its microenvironment in the hostile acidic environment of the stomach causing mucosal inflammation, in some patients and increases the severity of peptic ulcer disease.
Nowadays there is a well-established relation between the H. pylori and gastritis, duodenal ulceration. Gastrin and pepsinogen which are high in the infected patients and the somatostatin is low [22].
H. pylori produce virulent factors such as urease, catalase, vacuolating cytotoxin and lipopolysaccrides. Many of the patients with duodenal ulcers have impairment in the secretion of the bicarbonate in the duodenum and the causative agent is the H. pylori [23].
Increased gastric acid secretion and reduced duodenal bicarbonate secretion promote the development of gastric metaplasia i.e. gastric epithelium present in the first portion of the duodenum and this will cause duodenitis and predispose the duodenum to acids causing duodenal ulcer [22,24].
Etiology
Peptic ulcer disease (PUD) may be caused by:
• H. pylori infection which causes more than 90% of duodenal ulcers and up to 80% of gastric ulcers [25].
• Drugs (NSAIDs).
• Lifestyle factors, smoking is one of the causative agents of duodenal ulcer and can increase the relapse of PUD [26-28].
• Severe physiologic stress [22].
• Hyper secretory states (uncommon) [22].
• Genetic factors, 20% of patients are having a family history of duodenal ulcers [22].
H. pylori infection
H. pylori infection and Non-Steroidal Anti-Inflammatory Drug (NSAID) use constitute for most cases of PUD. The rates of occurrence were lower in whites than in non-whites. Prevalence of H. pylori infection in complicated ulcers (i.e., bleeding, perforation) is very low than that found in uncomplicated ulcer disease [22].
Drugs
NSAID use is very commonly a cause of PUD. It disrupts the mucosal permeability barrier, rendering the mucosa vulnerable to injury. Around 30% of adults taking NSAIDs have GI adverse effects. Other factors associated with an increased risk of duodenal ulcers in the setting of NSAID use include history of previous peptic ulcer disease, advanced age, female sex, high doses or combinations of NSAIDs, long-term NSAID use, concomitant use of anticoagulants, and severe comorbid illnesses [22]. Almost evidence now supports the assertion that H. pylori and NSAIDs are synergistic with respect to the development of peptic ulcer disease [22]. Regarding, Corticosteroids alone they do not increase the risk for PUD; although, they can potentiate ulcer risk in patients who use NSAIDs concurrently [22].
Epidemiology
Around two thirds of the world’s populations are infected with H. pylori. International statistics about the frequency of PUD is variable and mostly the causative agents are the H. pylori and NSAIDS where in United States, about 25 million Americans suffer from peptic ulcer disease, the annual incidence is 500,000 to 850,000 [25].
It is among the top 25 leading causes of death according to the World Life Expectancy Bangladesh profile data [29].
In Africa, studies from Morocco and Ethiopia have reported a prevalence of H. pylori infection of 75.5% and 65.7% respectively. In both studies they found a significant increase with age [30,31]. In Nigeria, they reported in one study, high prevalence of 80% when tested with histology and were even higher by 93.6% when serology was applied [32].
The prevalence of PUD is changed were the occurrence of the disease is similar in male and females it whereas was predominant in males [33,34]. It is more prevalent among older people, African Americans, Hispanic and in low socioeconomic group of population [25].
Diagnosis
The diagnostic method for the H. pylori related PUD is by the upper GIT endoscopy, where the biopsy urease test is done and this is a rapid testing can be performed when the biopsy is taken [14,35]. Barium swallow procedure is not commonly used only in some patients.
H. pylori testing- whenever peptic ulcer disease is confirmed H. pylori is tested by either taken a biopsy from the affected patient or by a urea breath test (C13, C14) and a stool test (Faecal antigen test sample). Also, blood test may be available but less reliable. PCR allows determination of antibiotic sensitivity and it has an excellent sensitivity and specificity but their methodology among labs are variable [36].
Treatment
Treating H. pylori infection is a challenge nowadays because of many reasons. The single important one remains the antibiotic resistance to the H. pylori [37].
Triple therapy with proton pump inhibitors (PPI regimen) was the mainstay one for treating H. pylori but now this line has developed a resistance for many reasons.
First is the duration of therapy, polymorphism of CYP2C19 for PPI and their rapid metabolism and the insufficient control of gastric pH and the consequent reduction in the antibiotic efficacy [37].
First line triple eradication regimen: Consist of Proton Pump Inhibitor (PPI) with two antibiotics given BID for 7 d.
Lansoprazole 30 mg po BID, metronidazole 500 mg po BID, clarithromycin 250 mg po BID [38]. Common side effects of the triple regimen: Diarrhoea, nausea [39] (Table 2).
| PPI based triple therapy | Esomeprazole 20 mg twice daily, OR omeprazole 20 mg twice daily. Amoxicillin 1 g twice daily. Clarithromycin 500 mg twice daily. | |
| PPI base Quadruple Therapy (E.TG, et al.) | Omeprazole 20 mg once daily. Bismuth subsalicylate 120 mg four times daily. Tetracycline 500 mg four times daily. Metronidazole (250-500 mg) TID or QID (500 mg) | Uncommonly used. Prescribed as a 7-14-d course. First choice line with known clarithromycin resistance [40]. |
Table 2. Currently recommended eradication regimens.
The Japanese Society of Gastroenterology (JSGE) revised the evidence-based clinical practice guidelines for peptic ulcer disease in 2014 and they concluded that if first-line eradication therapy fails, second-line therapy is given. In cases of non-H. pylori, non-NSAID ulcers or H. pylori-positive patients with no indication for eradication therapy, non-eradication therapy is given. The first choice is PPI therapy, and then the second choice is histamine 2-receptor antagonist therapy. After initial therapy, maintenance therapy is provided to prevent ulcer from relapsing [41].
Evidence for newer therapies
Sequential therapy: Due to the increased first line H. pylori infection therapy growing resistance to antibiotics used in this regimen [40]. The novel line treatment is sequential therapy which consists of two steps: PPI with amoxicillin for the first 5 d followed by triple therapy that includes a PPI, clarithromycin and tinidazole for another 5 d and this is a 10-d program.
An example regimen esomeprazole 20 mg twice daily combined with amoxicillin 1 g twice daily, prescribed for 5 d. This is followed by a triple therapy of esomeprazole 20 mg twice daily, clarithromycin 500 mg twice daily and tinidazole 500 mg twice for the following 5 d [42]. Common side effects to this therapy taste disturbances and diarrhoea [39].
The sequential treatment yielded superior eradication rates compared to standard therapy, 87.0% and 82.3% respectively [43].
Salvage therapy: Some patients fail to respond to the sequential therapy, for these patients’ salvage therapy is a triple therapy consisting of PPI, amoxicillin and levofloxacillin administered for 10 d. An example prescription includes esomeprazole 20 mg twice daily, amoxicillin 500 mg twice daily and levofloxacin 500 mg BID [44].
The mean intragastric pH values are high in poor metabolizers (4.1-4.47) whereas in intermediate (3.3) and rapid metabolizers (2.1-2.3), after a single 20 mg dose of omeprazole. Therefore, the standard recommended doses of any of these PPIs might not provide sufficient acid suppression in rapid metabolizers, so that increasing the dose might be required in this group [45].
Proton pump inhibitors
PPIs (e.g. omeprazole, lansoprazole) are responsible of inhibition of CYP2C19, which may cause drug-drug interactions when co-administered CYP2C19-metabolized drugs [7,46].
By reviewing the literature, it was shown that Proton Pump Inhibitors (PPIs) are extensively metabolized in the liver by CYP2C19, that demonstrates genetic polymorphism with 21 mutant alleles. Intragastric pH values and the plasma concentration of PPIs after oral ingestion were significantly lower in EMs named as normal homozygotes (CYP2C19*1/*1) and heterozygotes (CYP2C19*1/*X) compared to PMs named as mutant homozygotes (CYP2C19*X/*X) where 'X' represents the mutant allele. However, association has been found between the genetic polymorphism of CYP2C19 and therapeutic response to PPIs. CYP2C19 polymorphism affected eradication of Helicobacter pylori where using different PPI based eradication therapies as PM patients demonstrated significantly higher eradication rates compared to EMs [47]. One study reported that there is a successful eradication of H. pylori when using omeprazole in all patients who had little or no CYP2C19 activity, but in only 29% of patients who had normal CYP2C19 activity [48]. The longterm use of PPIs is associated with several adverse effects. Daily treatment with any PPI for longer than 3 y patient may develop mal-absorption of vitamin B12, caused by hypochlorhydria. The prolonged hypochlorhydria increases the risk of clostridium difficile infection, and also increase the risk for osteoporosis-related fractures, the FDA recommends that patients must use the lowest dose and shortest duration of PPI therapy appropriate to the disease being treated [49].
6 PPIs are FDA approved for clinical use esomeprazole, dexlansoprazole, lansoprazole, omeprazole, pantoprazole, and rabeprazole. All are of similar potency at inhibiting gastric acid secretion and are thought to be similarly efficacious [50]. Omeprazole is a Proton Pump Inhibitor its main action in the stomach is the blockage of gastric acid secretion. It is used for treating gastro oesophageal reflux disease (GERD), gastric ulcers, duodenal ulcers, and other acid related disorders. It is used also in the treatment of hyper secretory conditions, such as Zollinger Ellison syndrome and is used to eradicate H. pylori infection in combination with antibiotics.
Omeprazole: It was the first PPI to be introduced to the US market in 1989 [51]. Hydrogen ions (H+) are secreted actively in to the gastric lumen in exchange for potassium ions (K+) via an H+/K+-ATPase, this is also known as a “proton pump”. Located on the surface of gastric parietal cells, the proton pump controls the final step in acid secretion, and by targeting this step, omeprazole and the other PPIs are able to inhibit gastric acid secretion. Omeprazole is metabolized and inactivated in the liver by the CYP P450 system. CYP2C19 is the main enzyme involved, although other enzymes such as CYP3A4 may also contribute. Omeprazole is metabolized to two things hydroxy and des-methyl metabolites, which have no effect on gastric acid secretion [52]. Individuals with CYP2C19 enzyme activity reduction have a positive effect on clinical outcomes especially if taken for less than 6 months without increased risk of omeprazole toxicity [53].
Cytochrome P450 pharmacogenetics in Africa
After surfing the data base in PubMed, it is found that Africa harbours a disproportionate burden of disease when taking into account the triple challenge caused by HIV/AIDS, tuberculosis (TB) and malaria, against a backdrop of an increasing burden of no communicable diseases. The problem is more than 80% of therapeutic drugs used in the management of these diseases/ conditions are metabolized by CYP enzymes that exhibit genetic polymorphisms [54]. A study done in 2013 showed that there is high degree of variability when comparing individuals of African origin to other ethnic groups and also the distribution of clinically relevant CYP450 alleles amongst the various African populations. This data exemplifies the need for in-depth and well-orchestrated molecular and pharmacological investigations of these populations in the future, so genome sequencing and association studies will be critical [55].
Gene CYP2C19: The cytochrome P450 (CYP) is a large and diverse group of enzymes that form the major system for metabolizing lipids, hormones, toxins, and drugs. The CYP genes are very polymorphic and can result in reduced, absent, or increased drug metabolism. CYP2C19 is expressed in the liver and, to a lesser extent, in the small intestine [56]. Constitutive expression of CYP2C19 is mediated by hepatic nuclear factors 4α (HNF4α, HNF4A) and 3γ (HNF3γ, FOXA3) [57-62], and transcriptional activation is via drug-responsive nuclear receptors CAR (NR1I3), PXR (NR1I2), and GRα (NR3C1) [63,64]. These are regulated by endogenous hormones and by drugs such as rifampicin [65]. In addition to rifampicin, human CYP2C19 can be induced by ritonavir, nelfinavir, hyperforin, St. John’s Wort, dexamethasone, and artemisinin [66]. Recently, in-vitro expression studies have recently shown that the GATA-4 (GATA4) transcription factor also upregulates CYP2C19 transcriptional activity by binding to two predicted GATA-specific promoter elements [67]. Also, decreased CYP2C19 activity among women using steroid oral contraceptives can result from transcriptional downregulation of CYP2C19 expression through binding of ligand-activated oestrogen receptor α to a specific oestrogen response element consensus half-site in the CYP2C19 promoter [19]. In reviewing the CYP2C19 gene maps onto chromosome 10 (10q24.1-q24.3) and encodes a 490-amino-acid protein. Polymorphisms of several genes are known to be associated with reduced enzyme activity.
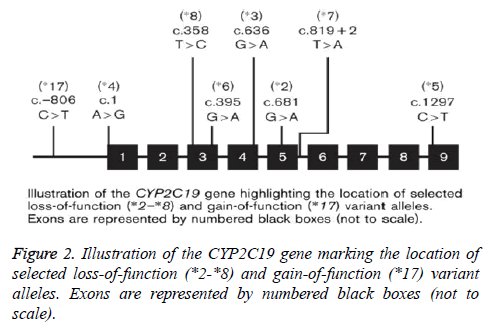
Recognizable among these are CYP2C19*2, characterized by a 681G-->A substitution in exon 5 leading to a splice-defective site, and CYP2C19*3, which has a point mutation in exon 4, which lead a premature stop codon. Therefore, individuals homozygous for the CYP2C19*2 and CYP2C19*3 alleles are considered to be Poor Metabolizers (PM), while subjects with at least one CYP2C19*1 allele are classified as extensive metabolizers (EM). More recent alleles identified include CYP2C19*17-21, few of which have been functionally characterized [16,68]. While the population prevalence of a number of CYP2C19 alleles, and their functional consequences, are well documented [69,70], this is less so for CYP2C19*17 [71]. So far 8 variant alleles (CYP2C19*2 to *8) have been identified which predict in a homozygous mutant or compound heterozygous manner the poor metabolizer phenotype. The variants CYP2C19*2, *3, *4, *5, *6, *7 and *8 are so-called null alleles with no CYP2C19 enzyme activity as consequence of a splice defect (*2), a premature stop codon (*3) or alteration of structure and/or stability of CYP2C19. However, recently several new CYP2C19 alleles have been identified (CYP2C19*9 to *15) in individuals from different racial groups with different ethnic background. At present, however, it is unclear whether these mutations are associated with significant alteration of in-vivo enzyme activity [72]. The CYP2C19 gene has nine exons and is highly polymorphic, with over 25 different variant stars (*) alleles currently identified by the Human Cytochrome P450 Allele Nomenclature Committee as shown in Figure 2.
Common variants which encode decreased or absent enzymatic activity
Polymorphism of the CYP2C19*2 allele is the defined rs4244285 (c.681G>A) and is a synonymous G>A transition in exon 5 that creates an aberrant splice site; this change the mRNA reading frame, which results in a truncated, nonfunctional protein [73]. CYP2C19*2 is the most common CYP2C19 loss-of-function allele, with allele frequencies of about 12% in Caucasians, 15% in African-Americans, and 29-35% in Asians [74,75]. Polymorphism of the CYP2C19*3allele is the rs4986893 (c.636G>A) defining and is a G>A transition in exon 4 that results in a premature termination codon at amino acid 212 (p. W212X) [76]. The CYP2C19*3 allele frequencies in most populations are below 1%; although, it is more prevalent in Asians (2-9%) [74]. Less frequent CYP2C19 alleles associated with absent or reduced enzyme activity are CYP2C19*4 (rs28399504), *5 (rs56337013), *6 (rs72552267), *7 (rs72558186), and *8 (rs41291556). These variants typically have allele frequencies less than 1% [74,77].
Drug metabolizer categories
According to the different combination of haplotypes an individual inherits in each drug metabolizing gene, a drug metabolizer status can often, but not always be predicted. There are 4 different drug metabolizer status types: poor metabolizers (also called “PM”), intermediate metabolizers (also called “IM”), extensive metabolizers (also called “EM”) and ultra-rapid metabolizers (also called “UM”).
Poor metabolizers: They have two non-functional alleles and therefore have little to no enzyme activity.
Intermediate metabolizers: they have one non-functional allele and one normally functioning allele, and though they have decreased enzyme activity.
Extensive metabolizers: They have 2 normally functioning alleles and therefore have normal enzyme activity.
Ultra-rapid metabolizers: Ultra-rapid metabolizers have one or more alleles which result in increased enzyme activity compared to extensive metabolizers.
The result of each metabolizer type on medication response depends on the role of the enzyme in the metabolism of the specific drug in question. For example, for a drug that is inactivated by the enzyme, an ultra-rapid metabolizer may need a higher dose of the drug to reach a therapeutic range while for another drug, that is activated by the enzyme; ultrarapid metabolizer status may be associated with increased exposure to the drug and therefore an increased risk of adverse drug reactions [70,78,79].
According to the ability to metabolize CYP2C19 substrates, individuals can be classified as Ultra-rapid Metabolizers (UM), Extensive Metabolizers (EM), Intermediate Metabolizers (IM), or Poor Metabolizers (PM). The wild-type gene is normally referred to as the *1 allele. For example, the CYP2C19*1 allele refers to the wild-type allele in the CYP2C19 subfamily [80]. EM individuals are homozygous for the CYP2C19*1 allele, which is associated with functional CYP2C19-mediated metabolism. The IM genotype consists of one wild-type allele and one variant allele that encodes decreased or absent enzyme function (e.g., *1/*2, *1/*3), result in reduced CYP2C19 activity [77]. PM individuals have two loss-of-function alleles (e.g., *2/*2, *2/*3, *3/*3), resulting in markedly reduced or absent CYP2C19 activity [77,81]. Of note, some CYP2C19 literature uses a separate nomenclature system that includes ‘homozygous extensive metabolizers’ (e.g., *1/*1), sometimes also referred to as ‘rapid metabolizers’; ‘heterozygousextensive metabolizers’ (e.g., *1/*2); and ‘PM’ (e.g., *2/*2). In spite of the nomenclature system, the frequency of CYP2C19 PMs is approximately 2-5% in Caucasians and African- Americans, and approximately 15% in Asians [74]. Individuals that carry one or two *17 gain-of-function alleles (e.g., *1/*17, *17/*17) may be categorized as UMs. Moreover, the phenotypic consequences of a loss-of-function allele and a *17 compound heterozygous genotype (e.g., *2/*17) is currently unclear but may be in between the EM and IM phenotypes, and possibly may be dependent on the substrate [2,82]. An important caveat in translating genetic information into predicted metabolizer status category is that the CYP2C19*1 allele is defined by the absence of other variants. Whereas, genotyping assays that do not query all variation in the gene may misclassify some individuals; if all common variants (i.e., >1% allele frequency) are genotyped, misclassification error will be small. The CYP2C19 enzyme involve in the metabolism of many clinically important drugs, such as antidepressants, benzodiazepines, and some of the PPIs, including omeprazole. CYP2C19 is highly polymorphic, as 35 variant star (*) alleles are currently identified at the Human Cytochrome P450 (CYP) Allele Nomenclature. The CYP2C19*1 wild-type allele is linked with normal enzyme activity and the “extensive metabolizer” phenotype [83]. The most common loss-of-function variant is CYP2C19*2, which consist of a c.681G>A variant in exon 5 that results in an aberrant splice site. This will lead to the production of a truncated and non-functioning protein. The CYP2C19*2 allele frequencies are ~15% in Caucasians and Africans, and ~29-35% in Asians [84]. “Intermediate metabolizers” they carry one copy of an allele that encodes reduced or absent function (e.g. *1/*2), whereas “poor metabolizers” were homozygous or compound heterozygous for two loss-offunction alleles (e.g., *2/*2, *2/*3). Also, commonly tested loss-of-function variant is CYP2C19*3, which contains a c. 636G>A variant in exon 4 that causes a premature stop codon. In CYP2C19*3 allele frequencies are ~2-9% in Asian populations, but rare in other racial groups. Another loss-offunction variant occur in less than 1% of the general population, and this include CYP2C19*4-*8 [85].
Eradication of H. pylori infection with omeprazole, lansoprazole, and pantoprazole has been found to be increased among CYP2C19IMs and PMs compared with Ems [2,3,86,87]. Additionally, the healing rates of peptic ulcers and gastroesophageal reflux disease during PPI treatment affected by CYP2C19 genotype [87] as IMs and PMs have been found to respond better to PPI treatment than Ems [2,3,87]. Regarding differences among PPIs are reflected in the different AUC values of rapid metabolizers compared with poor metabolizers. The AUC values for omeprazole and lansoprazole are 4 to 15-fold higher in poor metabolizers than in rapid metabolizers. The value for intermediate metabolizers is 2 to 3-fold higher than in rapid metabolizers. Pantoprazole is similar to omeprazole and lansoprazole; its AUC value of poor metabolizers is six-fold higher than in intermediate and rapid metabolizers [21].
Regarding the activity of CYP2C19*17 for most pharmacokinetic parameters of the substrates studied the average value was altered, the range of values showed mostly complete overlap for CYP2C19*1/*17 heterozygotes and wildtype homozygotes. Even for CYP2C19*17 homozygotes, the absolute effect was modest compared with the effect of previously identified loss-of-function alleles for example CYP2C19*2 and, CYP2C19*3. In Helicobacter pylori eradication CYP2C19*2 carriage was associated with an altered eradication rate (odds ratio 4.20, 95% confidence interval 1.23, 16.44) relative to the wild-type, but CYP2C19*17 homozygosity was not. Prevalence of the variant allele was typically<5% in Asians and about 4 times higher in White and African populations. CYP2C19*17 genotyping is unlikely to have clinical utility except for drugs with very narrow therapeutic indices [71].
Genetic testing
Pharmacokinetics of drugs and their therapeutic efficacy depend on the metabolic rate of activation or elimination. Poor metabolizers have markedly reduced or no conversion rate of pro-drugs, which eventually require alternative treatment or increased dosage. Poor metabolizers may have a reduced capacity to eliminate active drugs leading to accumulation and resulting in toxicity and life-threatening side effects. Such adverse effects can be predicted by determining the genotypephenotype associations. The homozygous wild type allele of CYP2C19, designated CYP2C19*1, and is defined as an extensive metabolizer (EM). Nine common variants of CYP2C19 gene is associated with impaired drug metabolism include *2 to *10. While the wild-type form of the CYP2C19 gene (*1) encodes an enzyme with normal functional activity, genetic variants (*2 to *8) encode enzymes with nonfunctional activity and two other variants (*9 and *10) have decreased functional activity. The most common Poor Metabolizer (PM) phenotypes are from CYP2C19 with homozygous *2 and *3 alleles. Homozygous *4 to *8 alleles are less common PM. One of the variants (*17) confers augmented enzyme activity, leading to an ultra metabolizer (UM) phenotype. Inheritance of one normal allele and one nonfunction allele can result in intermediate metabolizer (IM) phenotype. This genetic test is used for identification of patients at risk for adverse events due to impaired metabolism by: Detecting 6 variants for CYP2C19 gene responsible of nonfunctional metabolic activity: CYPC2C19*2, *3, *4, *6, *7, and *8 detecting 2 variants of CYP2C19 gene responsible of decreased functional metabolic activity: CYPC2C19*9 and *10 Detecting one variant of CYP2C19 gene responsible for increased functional metabolic activity: CYPC2C19*17 (72) Currently, the FDA does not provide recommendations about the use of CYP2C19 genetic testing for omeprazole treatment. Usually a patient’s result is reported as a diplotype, such as CYP2C19 *1/*1, and may include an interpretation of the patient’s predicted metabolizer phenotype (ultra-rapid, extensive, intermediate, or poor) [49]. However, a number of genotyping technologies can be used to interrogate variant CYP2C19 alleles in Clinical Laboratory Improvement Amendments-approved laboratories, two genotyping platforms have been approved by the US Food and Drug Administration at the time of this writing: the AmpliChip CYP450 Test (Roche Molecular Systems, Inc., Pleasanton, California, USA) that interrogates CYP2C19*2 and *3 (plus CYP2D6 variant alleles) and the Infiniti CYP2C19Assay (Auto Genomics, Inc., Vista, California, USA) that interrogates CYP2C19*2, *3, and *17 [74].
CYP2C19*1 is the wild-type allele and is found in individuals with normal enzyme activity. Individuals who are homozygous for the *1 allele are known as “extensive metabolizers” [88]. The commonest loss-of-function variant is CYP2C19*2 (c. 681G>A), were allele frequencies of ~15% in Caucasians and Africans, and 29-35% in Asians [89]. It is inherited as an autosomal co-dominant trait [74]. “Intermediate metabolizers” usually carry one copy of an allele that encodes reduced or absent function (e.g. *1/*2), whereas “poor metabolizers” are homozygous for two loss-of-function alleles (e.g. *2/*2). In Swiss a study was done and revealed new allelic variant that contribute to the PM phenotype in Caucasians CYP2C19*7 contained a single T-->A nucleotide transversion in the invariant GT at the 5’ donor splice site of intron 5 [90].
In reverse to that the non-functional alleles, the CYP2C19*17 allele (c.-806C>T) is associated with increased enzyme activity. Allele frequencies range from 3 to 21% in different populations [74]. Homozygous individuals for the *17 allele are known as “ultra-rapid metabolizers”, and it is this patient group who may benefit from an increased dose of Omeprazole. Although, not all studies have identified a significant effect of CYP2C19*17 on the metabolism of proton pump inhibitors and treatment outcomes [74].
In study done in China they identified the incidences of different CYP2C19 mutant allele (*2 and *3) frequencies in the Hakka population in southern China and they found mutations in this ethnic group. Alleles frequency in CYP2C19*2, and *3 varies among different ethnic groups as it constitutes about in Hakka population in China 0.3106 and 0.0461 [91].
Different alleles frequency in CYP2C19*2 and CYP2C19*3 in different ethnic groups in many countries in Asia, and Africa where 0.21 and 0.12 in Koreans, 0.30 and 0.11 in Japanese, 0.24 and 0.14 in Vietnamese, 0.23 and 0.05 in Malaysian, 0.12 and 0.00 in Turkish, 015 and 0.13 in Saudi Arabian, 0.24 and 0.20 in Iranian, 0.23 and 0.00 in Swedish, 0.18 and 0.01 in Tanzanian, 0.14 and 0.02 in Ethiopian, and 0.13, 0.00 in Zimbabwean [92].
The CYP2C19*2 allele occurs is frequently among Indian populations (North India, 33.1%; South India, 36.8%) than among Africans (16%), Caucasians (13.3%) and Asians (28.4%). The CYP2C19*3 allele in Indians is 1.9% in North India and 1.1% in South India. The of loss-of-function alleles (CYP2C19*2 and *3) varies greatly inter and intra-ethnic variations among Indian populations compared to other major ethnics as Asians (28.4% and 10.1%) and Caucasians (13.3% and 0.2%) respectively [93].
Significant interethnic differences exist in the incidence of PMs among Asians (13-23%), Africans (6%), and Caucasians (2-5%) [93].
CYP2C19*2 was found at various frequencies across all subpopulations studied, thus it constituted the predominant dysfunctional allele. The data also revealed the presence of *3 in Malays, Chinese, and Indians at low frequencies but it was not present in the Javanese from Indonesia, and Saudis from Saudi Arabia [94].
The genotypic polymorphism is effective on the rate of eradication of H. pylori. CYP2C19 genotype influences the eradication treatment rate with proton pump inhibitors like Rabeprazole [95].
Clinical CYP2C19 pharmacogenetic testing
However, a number of genotyping technologies can be used to interrogate variant CYP2C19 alleles in Clinical Laboratory Improvement Amendments-approved laboratories, two genotyping platforms have been approved by the US Food and Drug Administration: the AmpliChip CYP450 Test (Roche Molecular Systems, Inc. Pleasanton, California, USA) who interrogates CYP2C19*2 and *3 (plus CYP2D6 variant alleles) and the Infiniti CYP2C19 Assay (Auto Genomics, Inc., Vista, California, USA) which interrogates CYP2C19*2, *3, and *17 [45]. In 2016, Centre for Devices and Radiological Health, FDA approved Spartan RX CYP2C19 Test System by Spartan Bioscience manufacture *1, *2, *3, *17, DNA can be extracted by Buccal swab approximate turnaround 1 h [96,97].
Prognosis
With the appropriate eradication of H. pylori and the rational use of anti-secretory therapy and avoidance of NSAIDs decrease the recurring ulcer rate from 60-90% to about 10-20% [98].
The CYP2C19 dependent disposition and action of proton pump inhibitors showed that about 70% of the Caucasian populations (homozygous extensive metabolizers) will benefit from a higher dosage of the proton pump inhibitors that would increase Helicobacter pylori eradication in peptic ulcer and healing rates in gastro-oesophageal reflux disease by approximately 20%. Such therapeutic gain would necessitate genotyping of the patients for the common mutant alleles of CYP2C19 (e.g. CYP2C19*2 and eventually *3) before starting treatment with proton pump inhibitors as a predictive surrogate of the phenotype (e.g. AUC of proton pump inhibitors) [72].
Methodology
Study subjects
This was a prospective study. The study protocol approved by the principles of each of these hospitals; Fedail Hospital, Soba Hospital and Royal Care Hospital; In total 139, unrelated subjects (age 14-38 y) 48.2% (age 39-80 y) 58.1% who visited Fedail, Soba, and Royal Care Hospital, Khartoum, Sudan, were enrolled in the study from February 2016 to January 2017. Written informed consent was obtained from all participants prior to enrolment in the study.
DNA extractions
Saliva sample (1.5-2 ml) were collected in 25 ml sterile plastic containers from each participant and stored in the Deep freezer at -20°C. G-spinTM Total DNA extraction Mini Kit (iNtron Biotechnology, Inc. Cat #No 17046) G-spinTM Total Kit (Spin type), DNA concentration was quantified using the Nanodrop 2000TM Spectrophotometer (ThermoFisher Scientific, Waltham, MA) following the manufacturer’s instruction has been developed to extract high purity DNA, DNA references: 10.000-20.000 copies/μl. Only quality DNA (A260/280 ratio>1.7) was stored at -80°C until analysis.
Polymerase chain reaction
CYP2C19 genotype status was determined by Polymerase Chain Reaction (PCR)-restriction-fragment-length polymorphism. The single-nucleotide polymorphisms CYP2C19*2, CYP2C19*3 and CYP2C19*7 were genotyped using a commercially available kit (iNtron Biotechnology, Inc. Cat #No 17046). Polymerase chain reaction (PCR) was performed according to the following protocol: 50°C for 5 min, pre-denaturation at 94°C for 5 min, followed by 35 cycles of denaturing at 94°C for 25 s, annealing at 48°C for 40 s, and extension at 72°C for 30 s, and a final elongation at 72°C for 5 min.
Statistics
SPSS ver. 17 was used for data analysis Chi-square test and Ttest is used to correlate the allele and genotype frequencies and descriptive analysis was used to compare allele frequencies among different Sudanese ethnic groups. A value of P<0.05 was considered statistically significant.
Study design
Descriptive, cross sectional population based, in which selected Sudanese ethnic groups with peptic ulcer disease and treated with Omeprazole were used for the study.
Study area
Fedail Hospital, Soba Hospital and Royal Care Hospital in Khartoum City, Sudan.
Study duration of samples collection
Total duration of the research, February 2014-December 2017; duration of the sample collection is 6 months; duration of the sample analysis 4 months (2 months for PCR, 2 months SPSS).
Target population
Patients with peptic ulcer disease and treated with omeprazole.
Specific population characteristics
Patients diagnosed to have peptic ulcer disease and treated with omeprazole for 7-14 d from February 2016-January 2017 from selected Sudanese ethnic groups the Arabs, Nubians, Beja, Nuba, Darforians, Nilotics and Fulani.
Sample size
All patients presented in the period of study from February 2016 to January 2017 with peptic ulcer disease treated with omeprazole from different Sudanese ethnic groups are included in the research. The collected samples were from 139 patients of the selected group who satisfied the selection criteria.
Sampling method
A purposeful convenient sampling; in which patients with peptic ulcer disease and treated with Omeprazole will be reviewed and the PCR is done to differentiate the CYP2C19 different alleles in patients presenting from February 2016 to January 2017.
Study protocol
Ethical consent from Fedail and Soba Hospital and Royal Care Hospital was taken to take saliva sample from patients with peptic ulcer disease diagnosed by endoscopy or Urea breath test that take Omeprazole and for more than one-time treatment then consent is taken from each patient before interviewing or taking the saliva sample. Questionnaire is filled by the researcher himself, from each patient report information, regarding their gender, age, ethnic groups and the treatment received; all subjects were in good health based on medical history, physical examination, ECG evaluation and routine laboratory tests (blood chemistry, haematology, and urine analysis). The subjects were not taking any prescription or nonprescription medication 2 weeks before and throughout the study. Patient in outpatient clinic of gastro enterology endoscopy unit including the refer clinic of Dr. Suliman Salaeh Fedail and Dr. Nada Zakria in Fedail Hospital and Dr. Hatem Mudawi in Soba Hospital and Dr. Nada Mohamed in Royal Care Hospital are followed and selected according to the inclusion criteria.
Selection of patients
A total of 139 Sudanese patients were studied in the Endoscopy Units of different centres in the endoscopy units of Soba Hospital, the endoscopy unit of Professor Suliman Salih Fediel in Fedail Hospital and the endoscopy unit of Royal Care Hospital and all patients were either endoscopically or having their urease test positive for H. pylori and confirmed as having Peptic Ulcer disease.
All patients had omeprazole and they present again with peptic ulcer disease symptoms and take the eradication treatment more than once. The patients were from different ethnic groups.
Both sexes were included. The patients’ ages are 18 y and above. Patients on NSAIDS, patients with gastroduodenal inflammation due to systemic disease and patients who had received antibiotics in the last 2 weeks were excluded. The entire selected group they are diagnosed gastroduodenoscopy was repeated four weeks after completion of treatment to assess the healing of the gastroduodenal inflammation. Rapid urease test was performed on each biopsy separately. For confirmation of eradication of H. pylori, a single negative biopsy does not necessarily indicate eradication of the organism. If both the antrum and body biopsies were negative as assessed by the rapid urease test, the treatment will be considered successful and considered to be failed if both or either of the two biopsies was positive. After confirmation of the diagnosis and the omeprazole receiving by the patients more than once a saliva sample is collected in sterile containers and kept in deep freezer and then PCR is done to each sample
Inclusion criteria
Patients; males and females up to 80 y diagnosed by endoscopy or urease breath test to have peptic ulcer disease and treated with Omeprazole and there is a therapeutic failure is eligible to participate in the study.
Variables under study
Age, gender, ethnic group, treatment of peptic ulcer disease, duration of treatment, CYP2C19*2 allele, CYP2C19*3 allele, CYP2C19*7 allele.
Sample collection
Through regular visits to Fedail Hospital and Soba hospital refer clinic and attending upper GI endoscopy unit, a specifically designed questionnaire used and collected by the researcher from each selected participant. The researcher was available to identify the type of PPI used in the triple eradication therapy as many patients they do not know their medicine only by colour, by looking to the old prescriptions and seeing the tablets with them omeprazole has been identified.
Also, researcher makes sure that the patients received more than once omeprazole based triple eradication therapy and did not respond when he is diagnosed by Urease breath test or endoscopy to have positive Helicobacter urease test positive. Then saliva 1.5-2 ml (BioMed Central BMC, Nunes et al.) is collected in 25 ml sterile plastic containers from the selected group preserved in the refrigerator of the hospital in the endoscopy unit till the collection of the day finish carried in freezer flask to be transported to the Deep freezer at -20°C.
Extraction of genomic DNA only takes 20 min~30 min. Optimized protocol is offered for various kinds of samples’ I (Sib Enzyme Ltd Russia Cat No. E091) is used, SP-Taq DNA polymerase-specially Purified Taq DNA polymerase for PCR diagnostics. Also, i-Taq (TM) DNA Polymerase CAT 25021 (250 Units). Taq DNA polymerase optimally compose DNA at 72, therefore it could stably amplify a specified oligo sequence without adding enzyme at every cycle due to its thermal stability even at 94. i-Taq DNA polymerase consists of a single polypeptide chain with a molecular weight of approximately 95 kD. It is a highly processive 5'-3' DNA polymerase lacking 3'-5' exonuclease activity. i-Taq (TM) DNA polymerase, PCR buffer (w/15 mM MgCl2), MgCl2 free PCR buffer, 10 mM dNTPs, and 25 mM MgCl2 Unit: 5 U/li-Taq (TM) DNA polymerase
Unit definition: One unit is defined as the amount of enzyme required to catalyse the incorporation of 10 nmol of dNTP into an acid-insoluble form in 30 min at 74°C. The reaction conditions are in upper PCR working buffer, and 200 M dNTPs, and radiolabelled dTTP, and 150 g/ml activated calf thymus DNA.
Stability: Store at -20°C, and then stable for over 1 y.
Data analysis
Data analysis will be done using SPSS ver. 17, in order to fulfil the objectives of the study. Correlation test is done to detect the relationship between numerical variables, Chi square test is done to determine the significance of correlation between categorical variables, and T test used for establishing correlation between a numerical versus the categorical variables. Significance level will be taken for values equal to or less than 0.05.
Ethical consideration
Scientific and ethical approval from University of Medical Science and technology, Department of Pharmacology administration upon the proposal submission; participant’s confidentiality has been guaranteed. After talking to the patient and clearly discussing with him the specimen and the research objectives and the ethical consent is taking from participant. After talking to the patient and clearly discussing with him the specimen and the research objectives and the ethical consent is taking from participant.
Methods
Sample size: 139 saliva samples (Table 3).
| Urease C gene for H. pylori | Positive | Negative |
|---|---|---|
| Patient number | 127 (139) | 12 (139) |
Table 3. PCR for Detecting of urease C gene of H. pylori.
DNA extraction Method by using G-spinTM Total DNA Extraction Mini Kit (iNtRON Biotechnology, Inc. Cat# No: 17046).
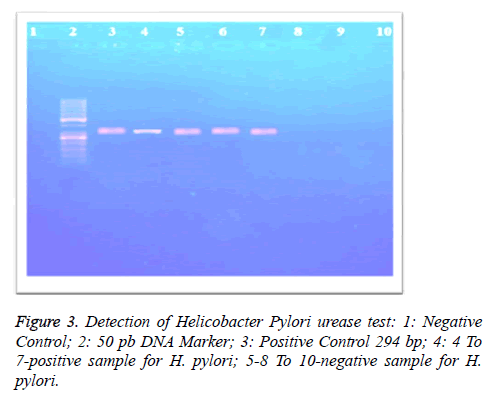
Primer using: 5`-AAG CTT TTA GGG TTGTTA GGG GTT T-`3 and for the Reverse primer was 5`-AAG CTT ACT TTC TAA CAC TAA CGC-`3. Master mix was prepared to reach a total volume of 20 μl: (5.0 μl 10X reaction buffer, 5.0 μl dNTPs 10 mM, primer set final 100 nM each (forward and reverse primers), 0.5 μl Prime TaqTM DNA polymerase (i- TaqTM DNA Polymerase/Cat. No. 25021) (iNtRON Biotechnology, Inc., Korea), positive PCR control (Amplirun® Helicobacter pylori DNA Control (Vircell S.L. Spain Cat No. MBC049), 5.0 μl DNA, RNAse-free water (Figure 3).
The thermal cycler was programmed in three steps: (Step l: 5 min at 94°C; Step 2: 30 s at 94°C, 30 s at 52°C. 1 min at 72°C, this was repeated for 35 cycles; Step 3: 5 min at 72°C) (Table 4).