Review Article - Journal of RNA and Genomics (2019) Volume 15, Issue 1
A Review on Biomedical Mining
Kosaraju Chaitanya, Rachakonda Venkatesh and Thulasi Bikku*Department of CSE, Vignan’s Nirula Institute of Technology and Science for Women, India
- *Corresponding Author:
- Thulasi Bikku
Department of CSE
Vignan’s Nirula Institute of Technology and Science for Women, India
Tel: +91-9838115336
E-mail: thulasi.jntuk@gmail.com
Received Date: 30 July 2019; Accepted Date: 15 November 2019; Published Date: 22 November 2019
© Copyright: Kosaraju Chaitanya, Rachakonda Venkatesh and Thulasi Bikku. First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Abstract
A wide range of biomedical repositories is available in the distributed systems for clinical decision making. One of the most important biomedical repositories is PubMed, which gives access to more than 50 million documents from MEDLINE. Data mining is used to explore hidden and unknown patterns from the large databases. The unstructured and uncertainty problems are available for many domain fields such as biomedical repositories, biomedical databases, web mining, health care system, education and technology intensive companies due to its large size. Information extraction from biomedical repositories and analyzing this information with an experimental study is time-consuming and requires an efficient feature selection and classification models. The latest trends of text mining are able to answer many different research queries, ranging from the biomarkers, gene discovery, gene-disease prediction and drug discovery from biomedical repositories. As a result, text mining has evolved in the field of biomedical systems where text mining techniques and machine learning models are integrated using high computational resources. The initial contribution of this research is to discuss the background study about biomedical repositories, document classification, clustering the relevant documents based on MeSH terms, methodologies, and models used in Biomedical Document analysis using the Hadoop framework. The main purpose of the work is to explain about the importance of feature extraction and document classification models to find gene-disease patterns from the massive biomedical repositories.
Keywords
Biomedical data analytics, Hadoop framework, Document Clustering, Document Classification, Parser.
Introduction
In the field of bioinformatics, collecting and searching the publications or documents plays a key role, due to their unstructured format of data and they are not grouped according to the keywords. From the past few decades the data has been increased exponentially in the field of bioinformatics, so it is a difficult task for a user to search the relevant data based on the user criteria for decision making. In this paper we discuss about traditional data mining extraction to latest document extraction and analysis. In the bioinformatics an ecosystem that transforms case-based studies to large-scale, data-driven research in big data [1].
The challenges of bioinformatics are storing, managing, and analyzing massive amounts of medical datasets. The automatic classification of medical documents into predefined classes is growing rapidly on online data repositories, one of the biggest problems motivated to assist experts in finding useful information from a large amount of distributed document repositories [2]. In distributed biomedical systems, text classification models are important as it can lead to advances in decision making including gene functions, gene-disease patterns, gene-gene associations and Medical Subject Heading (MeSH) knowledge discovery [3]. It is important to classify and organize the biomedical databases so users can access the useful information easily and quickly. As of late, the quantity of information sources in social insurance industry has developed quickly because of broad utilization of portable and wearable sensors innovations, which has overwhelmed human services territory with a tremendous measure of information. Hence, it winds up testing to perform medicinal services information examination dependent on conventional strategies which are unfit to deal with the high volume of enhanced medical information [4]. In general, healthcare domain has four categories of analytics: descriptive, diagnostic, predictive, and prescriptive analytics; a brief description of each one of them is given below.
Descriptive analytics
It consists of describing current situations and reporting on them. Several techniques are employed to perform this level of analytics. For instance, descriptive statistics tools It consists of describing current situations and reporting on them. Several techniques are employed to perform this level of analytics. For instance, descriptive statistics tools
Diagnostic analytics
It aims to explain why certain events occurred and what the factors that triggered them are. For example, diagnostic analysis attempts to understand the reasons behind the regular re admission of some patients by using several methods such as clustering and decision trees [5].
Predictive analytics
It reflects the ability to predict future events; it also helps in identifying trends and determining probabilities of uncertain outcomes. An illustration of its role is to predict whether a patient can get complications or not. Predictive models are often built using machine learning techniques.
Prescriptive analytics
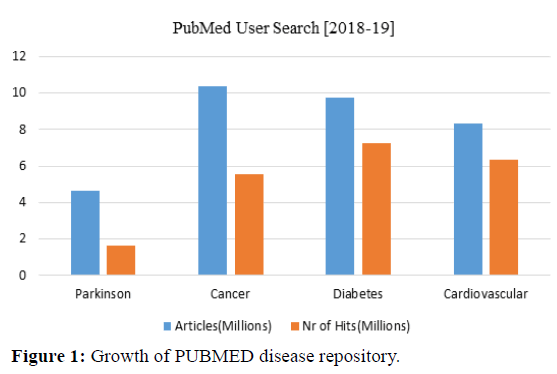
Its goal is to propose suitable actions leading to optimal decision-making. For instance, prescriptive analysis may suggest rejecting a given treatment in the case of a harming side effect high probability [6]. Figure 1 illustrates analytics about four diseases for the search results by using the tag name drugs, genes, diseases and medical terms on the PubMed disease repository.
In biomedical research, big data frequently contains an assortment of datasets from different information sources like Medline/Pubmed, Epigenomics, PROMIS, EyeGENE etc including authorized, randomized or non-randomized clinical investigations, distributed or unpublished information, and medicinal services databases [7]. Here the search results are shown in reverse chronological order, i.e., it shows documents according to the date and time or frequently accessed list. Boolean operators were integrated along with MeSH (Medical Subject Heading) terms for document retrieval according to the query construction, MeSH deals with the real content of articles [8]. MeSH database is responsible for finding and choosing MeSH terms, check the definition and document entity information make PubMed search strategy, show MeSH hierarchy, associate sub-headings and establish a link to MeSH browser [9].
In the Evidence-based biomedical disease forecasting methodology, involves retrieving relevant medical documents from PubMed databases by analyzing the user’s history of navigation and documents relevant to gene or protein structure. In PMC archive, there are almost 3.1 million Articles available, which was designed and developed by the National Institute of Health’s National Library of Medicine (NIHINLM). Each document is assigned a Unique Article Identity document (UAID). All the articles are stored in the XML document format are available publicly as well as freely, which is maintained by BioMed Central (BMC) [10]. BMC is defined as an open access journal publisher. Almost all BioMed Central journals are published online only. Genetic Association Database is an integration of human genetic association concepts related to critical diseases as well as disorders. Machine learning methods tend to be ineffective for large number of categories during the classification; MeSH contains more than 26k types of different categories. The original documents of biomedical repositories are in PDF or XML format are converted into ASCII files [11]. The integration of big data technologies in healthcare analytics may lead to better performance of medical systems.
Biomedical Document Analysis
Traditional distributed biomedical data mining, which provides scalability of data, enables to transform the high dimensional datasets into smaller datasets with an adequate computational resource to process the data effectively and efficiently [12]. Generating frequent item set is the more expensive because of the large data. Retrieving the frequent item sets is an essential part of all association rules mining strategy, where the association between the items is calculated to obtain the frequent item sets. The association relationships among a set of items in a dataset transaction is discovered using association rule mining [13]. There are some approaches which are responsible for discovery of multi-class association rules such as multi-class classification and multi-label associative classification [14]. The above approaches result enhanced accuracy as compared to the other conventional classification schemes. Another technique is developed which is an Apriori like algorithm is known as Associative classifier for negative rules [15]. Association rules may have positive and negative association rules, which take part in the process of constructing associative classifier. As compared with the positive rules of association, the negative association rules are more in number, which occupy more search space and used to construct negative association rules, can also be used to construct classifiers. With the rapid growth of evolution in mechanisms to detect frequent itemsets in transactional databases, association rule mining approaches also came into existence. The prime objective of association rule mining is to detect the correlation among datasets from different databases [16]. Traditional Data mining algorithms are used on a single large repository, which is static, but it is a challenging task for limited resources to process the data that is growing exponentially. To process these massive amounts of data, the software framework Map/Reduce is used to process the data parallel across the cluster of processors in Hadoop environment [17]. It is necessary to discover hidden knowledge or patterns from those databases to improve the decision making. Since, distributed data repositories are popularly categorized by privacy, heterogeneity and cross- platform; it is hard to perform the traditional data mining models. To overcome these issues, distributed data mining (DDM), acts as an extension of traditional data mining models in distributed environments on massive databases. The primary approach to the distributed data mining is that the data is uniformly distributed over a large number of distributed data repositories, process it and derive patterns through data mining techniques that reflect the features of the whole document set. The algorithms used on central large repository are converting to distributed environment using Hadoop Map/Reduce to improve the performance.
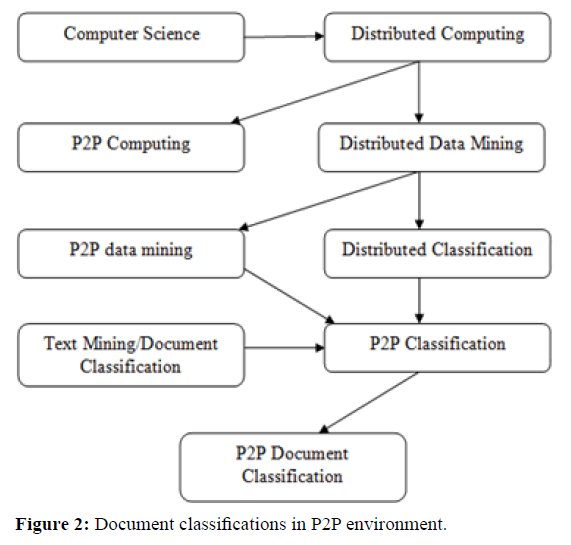
Figure 2 illustrates the various fields in distributed computing using data mining technologies. The task of the distributed document classification is to transform a large number of documents into relevant user-specific patterns. The main requirements for performing distributed data mining in large- scale systems are:
• To transmit the local domain knowledge to a centralized data center for feature extraction and document analysis.
• To develop a novel classification model on the centralized database, this is connected with a large number of distributed data centers.
• To combine the selected representatives from each local peer to the centralized peer node.
• To classify the document sets without a centralized functionality of the peer to peer model.
The task of the distributed document classification is to transform a large number of documents into relevant user- specific patterns. Conventional document classification methods have been implemented in the centralized databases with limited computational resources and data size. According to the divide-and-conquer method, a large problem is decomposed into smaller sub-problems, these sub-problems do not depend on each other and those sub-problems are solved in parallel by different mappers. After execution of all mapper, the outcomes are sorted lexicographically according to the respective output data key. Data values along with the same key are also sorted in the same machine and executed within the particular reduce task. Hadoop framework provides improved cluster utilization in distributed environment, resilient to failure, cost effective, highly scalable, supports novel programming models and services and agility [18].
Biomedical Data Analysis
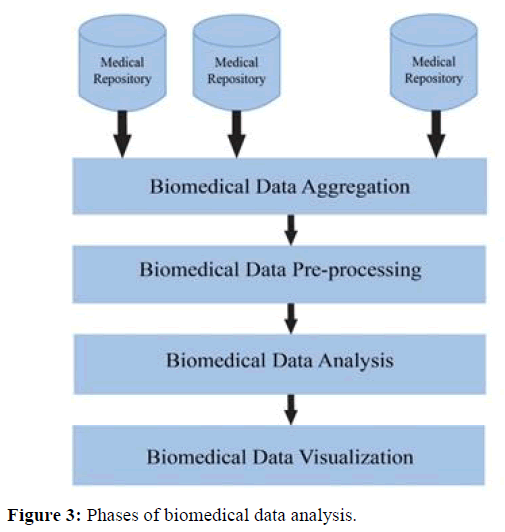
The preprocessing of the data needs to be done; the important and efficient features or attributes are extracted so as to reduce the curse of dimensionality. The data analysis is performed by using classification or clustering algorithms. The results are visualized as Graphs, Statistical graphics, plots etc. based on the requirement of the query. The biomedical text documents are tokenized using Part-of-Speech Tagging or in a bag-of-words approach like word stemming which removes prefix and suffix of a word and English stop words like full stop, comma, semicolon, colon etc. are filtered [19]. The complete analysis of the biomedical data is shown in the Figure 3.
Biomedical document pre-processing
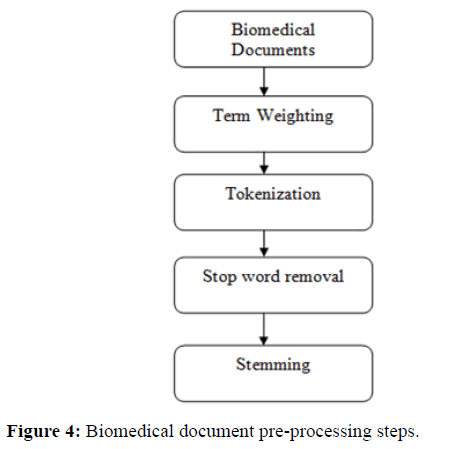
Many document preprocessing techniques have been implemented in the literature on biomedical repositories, which are responsible for transforming the raw information into a specific structured format. With the huge amount of digital data available in biomedical repositories, it has become important to implement a different text mining model that could efficiently control and manage the medical repository systems. There are two types of preprocessing process, i.e. abstractive and extractive preprocessing. Abstractive preprocessing converts original source document data into meaning sentences or phrases using linguistic methods. This technique is computationally expensive, hard to process on unstructured databases. On the other hand, Extractive preprocessing is a kind of summarizer which selects the phrases or sentences having the highest rank and organizes them in the central peer node for data analysis [20]. In the extraction phase, the weight of the phrase or sentence is computed using probabilistic weighted models. The weight of the sentence depends on statistical significance metrics, the presence of particular terms/ phrases and the position of the sentence. The main objective is to improve the quality of document features and minimizes the computation time in pattern mining as shown in Figure 4.
The basic steps in the document pre-processing are described as below:
Tokenization: Tokenization is the process of separating the document text into basic units known as terms or phrases. Biomedical raw texts are pre-processed and segmented into terms or phrases. The data must be operated in the three main steps for document tokenization process: the first step is to convert each document to term frequency which is known as Bag of Words (BOW). Almost all remaining tokens are words having a meaningful text format as described in WordNet (http://wordnet.princeton.edu/) or Unified Medical Language System (UMLS) (http://www.nlm.nih.gov/research/umls/). The chances of spelling mistakes are very rare in the case of Michigan Pain Consultant (MPC) dictations, as this approach involves an optimized quality transcription service [21]. The smaller tokens having low frequency cannot be discarded unnecessarily because of their low frequency. It provides outcomes having low weights in case of capture probability evaluation. Most of the biomedical communities have implemented Semantic Web technologies, including the construction of ontology, information extraction as well as knowledge discovery. Tokenization is the initial phase of document pre-processing; all the words acceptable by pattern matching algorithms are retrieved from various documents. There are several common words which do not affect the pattern extraction process are identified and discarded. Stop- word is defined as the most frequently used words which do not influence the pattern mining process. Some examples of stop- words are- delimiters, pronouns, prepositions, conjunctions, and so on. If the numbers of stop-words are decreased, the pattern mining process is enhanced to a great extent.
Stemming: Starting from elimination of suffix elimination and producing the word stems comes under Stemming [22]. Adding suffix to the root word like jump, jumped and jumping, here jump is the root word, the suffixes like ed, ing are added, those words to be considered as same words. These types of words are to be considered as single word, considering the root word and added to the dictionary, so as to reduce the storage and processing time [23].
Pruning: The process of eliminating words which are used rarely or frequently used in the document is known as pruning. Here, MEDLINE abstracts are extracted based on gene term, which is useful in pattern mining. The gene detection in the pattern mining can be done as the documents based on genes are identified and extracted from MEDLINE database. Next document preprocessing approach is implemented, feature selection, ranking, clustering are done [24].
Biomedical document analysis tools
Chunking is a natural language processing method that attempts to represent the document in the partial tree structure format, which is used in the preprocessing of Documents. Chunker splits the document content into a group of terms that contains a grammatical part, like noun, verb, and preposition phrases. In the statistical approach, statistical machine learning methods are used to chunk the biomedical datasets to a great volume. In the rule-based approach, a set of regular expressions is used to chunk the documents without training dataset. Here to analyze we use the UCI machine learning repositories, a corpus of annotated abstracts taken from National Library of Medicine’s MEDLINE database and the Unstructured Information Management Architecture (UIMA) framework to integrate the chunking software and assess the performance of the different chunkers. Here we have 468 different biomedical data sets (www.nactem. ac.uk/genia/genia-corpus) [25].
GATE chunker (http://gate.ac.uk): General Architecture for Text Engineering (GATE) is a special kind of framework used for the development and deployment of software components using natural language processing. GATE framework typically supports an object-oriented class library and extended to solve biomedical problems. GATE provides an efficient data structure for language and gene annotations in biomedical repositories. This tool provides an easy way to discover the differences between the two terms, phrases or MeSH terms with similarity scoring.
Genia tagger (http://www-tsujii.is.s.u-tokyo.ac.jp/GENIA/ tagger): Genia Tagger is a simple integration of chunk tag, Part-of-Speech (POS) tag and named entity detection tool. It is developed for the preprocessing of biomedical text; MEDLINE document sets. The Genia Tagger implements a sliding window technique that is based on maximum entropy formulation. Tagger models are based on the GENIA Corpus, Wall Street Journal (WSJ) Corpus and PennBioIE corpus. It is not possible to use other corpora to train a model.
Lingpipe (http://alias-i.com/lingpipe): Lingpipe is stated as a suite of Java libraries for natural language processing. This tool provides various features such as named entity detection, POS tagging, grammatical correction, and so on. This chunker usually supports rule-based approach, dictionary-based approach as well as statistical chunking approach. An improved version of Lingpipe is statistical chunker which depends upon the core idea of hidden Markov model. The Lingpipe architecture is considered as a very simple solution to implement document analysis models in other systems like Unstructured Information Management Architecture (UIMA). It supports a training mode as well as numbers of precompiled approaches for various domains.
Yamcha (http://chasen.org/~taku/software/yamcha): It is a text chunker which can be customizable, basic, and open source. It provides a various natural language processing features such as named entity detection, POS tagging, and text chunking. This is mainly works on support vector machine (SVM) algorithm, which can be easily processed, trained and merged with different applications.
OpenNLP (http://opennlp.sourceforge.net): OpenNLP is identified as an organizational open source natural language processing toolkit, which depends upon a maximum entropy measure. OpenNLP UIMA wrapper is designed to improve the text preprocessing procedures on small data repositories. This wrapper decomposes OpenNLP package into small sub-packages, which is responsible for performing sentence detection, tokenization, POS tagging, chunking, named entity recognition, and so on.
Metamap (http://mmtx.nlm.nih.gov): National Library of Medicine developed MetaMap, which is a highly configurable program to identify concepts from the Unified Medical Language System (UMLS) Metathesaurus in biomedical document. In view of the SPECIALIST negligible responsibility parser texts are part into pieces and distinguished as a concept. The SPECIALIST parser depends on the thought of an exceptional arrangement of alleged obstruction words that demonstrate limits between phrases. These hindrance words make it conceivable to run MetaMap without a training data for this model.
OpenNLP performed best on both noun-phrase and verb-phrase state acknowledgment, nearly pursued by Genia Tagger and Yamcha. OpenNLP performed best on both thing expression and action word express state affirmation, almost sought after by Genia Tagger and Yamcha. As for ease of use, Lingpipe and OpenNLP scored best. Blend of the explanations of the diverse chunkers by a basic casting a ballot plot is a direct method to enhance chunking performance and permits to adjust accuracy and review of the consolidated framework shown in Table 1.
| 100 Errors Randomly Selected | ||||||
| Chunker | Noun Phrases | Verb Phrases | ||||
| Precision | Recall | F-Score | Precision | Recall | F-Score | |
| Gate | 68.9 | 76.8 | 75.3 | - | - | - |
| Genia Tagger | 77.3 | 84.3 | 83.8 | 89.2 | 89.6 | 89.8 |
| Ling Pipe | 81.2 | 85.2 | 84.2 | 86.3 | 87.2 | 87.3 |
| Metamap | 79.6 | 86.2 | 86.01 | 69.4 | 75.3 | 69.8 |
| OpenNLP | 87.2 | 88.3 | 87.1 | 91.2 | 90.3 | 90.2 |
| Yamacha | 85.3 | 87.2 | 86.2 | 90.2 | 89.9 | 89.8 |
Table 1: Performance of the chunkers with randomly selected errors.
Biomedical document feature extraction
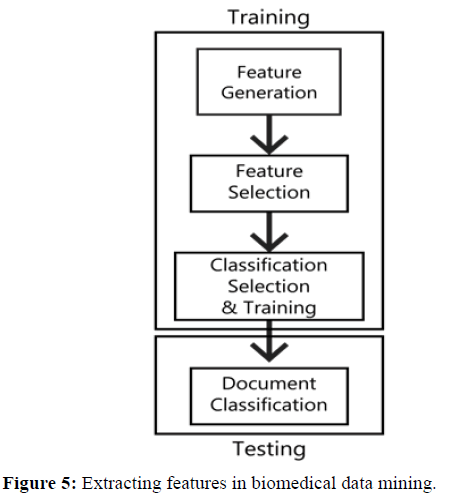
Classification also depends on the number of clusters, the centroid of the cluster and the type of domain or application. Cluster-based model mainly includes preprocessing task, classification and feature extraction. A multi-document feature extraction mechanism has been presented using a classification model which is based on pre-computed feature extraction that works for both single and multiple documents as shown in Figure 5. The feature extraction process has been introduced which contain three phases: Document Preprocessing, Soft Classification and Feature extraction. A model has been introduced for multi-document feature extraction by integrating document classification and feature extraction methods.
Be that as it may, the ongoing increment of dimensionality of information represents an extreme test to many feature selection and feature extraction techniques concerning proficiency and adequacy. In this era, where electronic textual data are increasing exponentially, and it is practically impossible for any user to read large volumes of individual documents. It is necessary to find strategies for permitting users to locate important information quickly within the collections of documents. Document features are represented in terms of sentences or phrases from different sources without any domain knowledge and thus making the information retrieval completely unbiased. Feature extraction is a highly interdisciplinary field in different domain fields such as information extraction, text mining, information retrieval, natural language processing (NLP) and medical databases. There are three types of methods to extract features from large datasets, they are Filter method, Wrapper method and embedded method [26]. The Filter method appears to be less optimal but executes faster than wrapper method. The results of this method are more general than wrapper method. This method is independent of classification algorithm, so the computational cost is very less for large datasets. The second method is wrapper method which is a best solution for supervised learning methods. This method depends on the classification algorithm, so computational is larger filter method, but gives accurate results than filter method. The last method is embedded method, which depends on classification algorithm with less computational cost and least prone to over fitting. It performs faster than wrapper method, but the performance is degraded when the irrelevant or duplicate features are more in target dataset. The main problems in bioinformatics area are: large dimensionality and small sample size. The multivariate selection algorithm for features is considered as one of the best algorithms.
Modern day approaches to feature extraction, try a variety of methods that attempt to handle the more sophisticated documents. One of the most recent works is the application of Non-negative Matrix Factorization (NMF) to feature extraction. This method focuses on the subtopics of a document like supervised latent dirichlet allocation (sLDA) and claims to be quite successful so it is applied in other related tasks to feature extraction, just like Latent Dirichlet Allocation (LDA), it is never previously applied to this specific task. Many graph-based approaches have also been developed for automatic feature extraction. Most of the early work with graph-based feature extraction builds upon work done in other aspects of Natural Language Processing, and, Information Retrieval. The rationale behind applying this method to this field is that document feature extraction is an information retrieval task where extracting the most important sentences to include in the summary.
Biomedical document ranking
Document feature extraction has been focused on summarizing large document sets using graph-based algorithms which incorporate ranking models. Most of the graph-based models have same functionality which contains preprocessing, execution model, rank-based algorithm and finally filtered output. A graph- based model is implemented TextRank which consider not only the local vertex information, but also extracts information from the entire graph recursively [27]. Steps involved in generating the Document Preprocessing are:
1. Identify vertices or nodes in the graph model as a phrase or sentence units which describe the given context for designing the graph model.
2. Based on the rank similarity measure model, add links in between the phrases or sentence and compute the rank similarity of each edge.
3. This graph model has weighted, or unweighted nodes or edges can be represented in the form of directed or undirected way.
4. Apply phrase or sentence ranking method in the graph model until all nodes are converged.
5. Compute a rank score for each node, based on final rank measures of each node in the graph, all the nodes are sorted for topic selected.
The model has been presented which uses TextRank with some differences and uses the shortest path method to find the nearest feature sets to the TextRank. In the initial phase, graph model has been built for representing the document and interconnected phrase entities in the graph model with meaningful relationships. A weighted graph method has been proposed using the novel approach which includes ranking both phrases and sentence classification for document feature extraction [28].
Major steps involved in this methodology are:
• Combines both sentence and phrase classification methods for similarity ranking.
• A phrase or sentence clusters are generated based on singular matrix factorization.
• The weighted graph model is implemented to find the sentence relationship in the documents.
This method has been presented in three phases. In the initial step, document structure is represented to every document in the document set; the structure can be represented as an un-directed graph. Phrases in the document play a significant role in the sentence formation in the graph model. In the second step, each phrase ranking measure in the document is computed using the ranking technique. Finally, the maximal marginal relevance technique is used to generate the relevant summary.
Biomedical document classification models
Biomedical Document classification has become an interesting research field. Partly, this is due to the increasing availability of biomedical information in digital form which is necessary to catalogue and organize [29]. In any case, past research has generally centered around semantically distinguishing biological entities like synthetic substances, ailments, genes and proteins with little exertion on finding semantic relations [30]. Document pattern mining automatically detects the similar documents using statistical measures on term frequencies, phrase frequencies, and sentence frequencies. The majority of the document pattern mining techniques are centered on the feature vector spaces, which are broadly used to train document model for text pattern mining. The similarity between sentences/documents is examined using one of document similarity measures that are based on such a feature vector or word frequencies, for instance, Jaccard measure and the cosine measure. Pattern mining techniques based on these vector spaces make use of single word i.e., one-gram interpretation only.
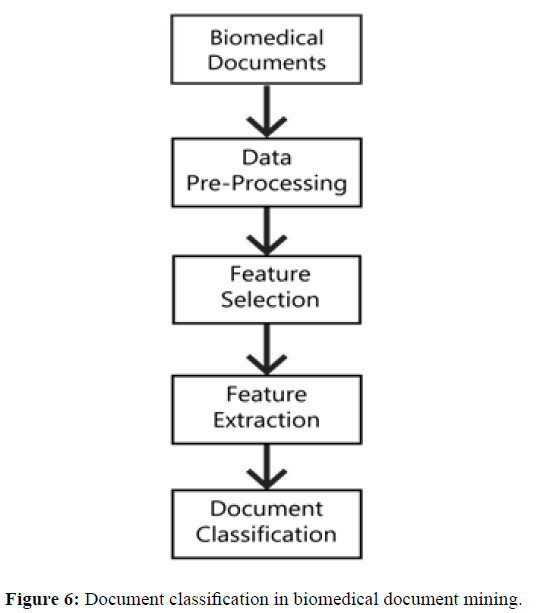
Document classification method is used to classify the high dimensional features for pattern discovery models which can be implemented in Hadoop environment as shown in Figure 6. Keyphrase extraction can be carried out to a single document for tagging the document/sentence. A consolidated document- based pattern mining can be labeled and filtered using key phrase feature extraction. Extended document patterns or classifications in flat biomedical repositories can be optimally filtered. Document pattern features can be interchanged between data centers to assist cooperative pattern mining. Distributed pattern mining in a hierarchical structure can be filtered level by level using specified threshold. The computational cost of the distributed pattern mining is very large, so the classification models are introduced to reduce the complexity in pattern mining [31].
Artificial Neural Networks (ANN) is recognized statistical learning algorithms dependent on neural systems. The neural network can be characterized as a system of neurons in charge of perceiving instances when invigorated. Learning should be possible via looking as indicated by system loads, which acknowledges obscure inputs for assessment and estimation of functions. KNN is also known as lazy learning, as there are no training phase and density estimation in the learning process. The kernel function of the KNN classification model optimizes the feature extraction of the biomedical documents. A definitive objective of this examination is to create machine learning classifiers that could decrease the manual effort important to audit noisy and missing accumulations of specific disease data. The overview of biomedical document classification models is discussed in the Table 2.
| Classification Model | Imbalanced Property/ Skewed Data | Training Data | Advantages | Disadvantages |
|---|---|---|---|---|
| Decision Trees [24] | Affected | Adequate training data with features and labels are required to avoid over fitting problems. | Robust to noise data; and decision rules evaluation. | Prone to over-fitting, Performance issue under the imbalanced property. |
| Bayesian Models [25] | Affected | Required to find prior and posterior probabilities. | Robust to probabilistic predictions. | Requires domain expert for decision making, Computationally expensive. |
| Artificial Neural Networks [26] | Affected | Data required for training model. | Able to learn non-linear functions. Robust against errors. | Difficult to interpret results, Slow training and prediction process. |
| Support Vector Machines [27] | Affected | Data required for training model. | The best model for high-dimensional datasets with complex kernel functions. | Slow processing, Low performance under limited features. |
| Ensemble Models [28] | Affected | Data required for training model. | Best model for high-dimensional datasets with complex feature selection models | Fast processing, Low performance under imbalanced data and missing values. |
| Random Forests [29] | Affected | Data required for training model. | Best model for Regression and classification, Overfitting is not easy in this case | Large number of trees makes algorithm slower, More number of trees required for good predictor, which slows down the model. |
| Random Vector Functional Link (RVFL) [30] | Affected | Data required for training model. | Learning speed is very fast with simple structure and generalization performance is good Avoids curse of Dimensionality | This model seriously affected with low g-mean values |
Table 2: Overview of hadoop based biomedical document classification models.
Information Extraction (IE) in the biomedical domain is the extraction of associations between biological entities in document sets. The most interesting patterns that are extracted from biomedical repositories are: Protein-Protein Interactions (PPIs), gene -protein, gene-disease and functional protein annotations. A large number of standard biomedical repositories are used for text classification to improve the model efficiency. Therefore, an efficient distributed classification method is required to enhance the accuracy, precision, accuracy, recall or sensitivity, specificity, F1 score on large biomedical repositories [32]. The confusion matrix is used for finding correctness and accuracy of the classification model.
Document classification is a process of mapping the content of a document into one (or more) of a set of pre-defined labels.
Biomedical document clustering models
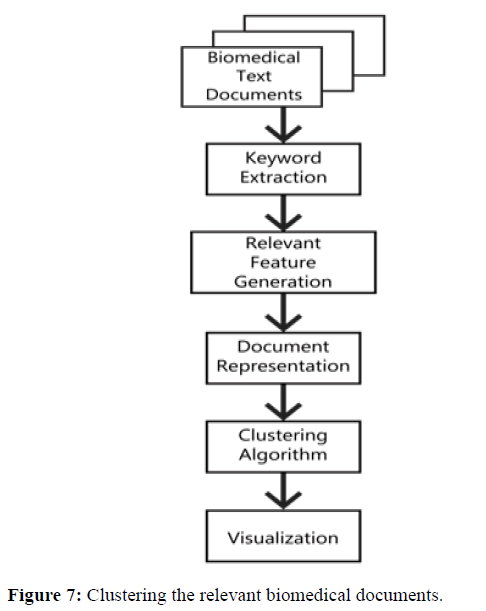
Clustering is a technique to group similar objects together based on their (dis)similarity, to form a grouping of objects such that objects in the same group are most similar while objects in different groups are more dissimilar. A potential benefit of clustering is to categorize the documents themselves. It might be possible to come up with groups of things that are recognized, but it might not be clear that they could be made into a category in advance as shown in Figure 7.
K-tree based document classification model is an improvement and an approximation of the k-means classification approach. Traditional k-means which represent clusters in a hierarchical manner, resolves the sparse representation problem in document representation model. They compare the quality and efficiency of the Classification Toolkit (CLUTO) algorithm using a set of documents [33]. The tree structure allows efficient space management in the main memory. The K-tree approach has been initially implemented on high dense vectors which minimize the error rate and space complexity problems [34].
According to some approaches, only a single outlier is determined and eliminated at a particular instance of time. The process continues until no more iterations or outliers [35] This can also give rise a serious issue of ignoring some important outliers. There are several other techniques which are capable of identifying multiple outliers simultaneously. The outlier detection techniques can be categorized into four types:
(1) statistical approaches, (2) distance-based approaches, (3) profiling methods and (4) model-based approaches. Statistical approach, the main limitation is unable to estimate the data point distribution of multidimensional data. In distance-based approach the dimensional distances of data points from one another using the available features are computed. In profiling approach, profiles of normal behavior are built using data mining techniques; the deviations are computed and considered it as intrusions. Finally, model-based approaches generally characterize the normal behavior data points using predictive model techniques and then detect outliers as the deviations from the normal learned model.
In the case of semi-supervised techniques, pre-labeled data are needed in order to determine the boundary of normality and enhances the process of classification for new data points. The new data point can either be normal or abnormal, which is evaluated by how these points are fitted in the normality model. In various real-world applications, it is very challenging to produce a set of representative normal data.
Conclusion
The research in Big Data is growing rapidly in all the domains and applications. Analytics models from these large data are expected to bring essential transformative and opportunities for different domain applications. Also, most of the traditional big data mining models and static classification models are not inherently scalable and efficient to find the essential hidden patterns on large distributed databases with high speed, high true positive, low error rate and incompleteness. This literature survey provides various methodologies for biomedical data to obtain the effective features from large data repositories to retrieve the knowledge according to users’ criteria in the Hadoop environment. The survey is used for the researchers and gives an idea how to use the necessary models and implementation of technologies required for their work, as well as for developers about how to provide more enhanced solutions for biomedical data analytics in support of decision making in the clinical and biomedical fields.
Acknowledgements
I thank my supervisor for his continuous suggestions and feedback; I thank our chairman Dr Lavu Rathaiah and Principal Dr Radhika Paturi for supporting me, who put their faith in me. This work is mainly based on biomedical data extraction from the corpus, so I thank the referees in this wide area. I thank each and every one who supported me to accomplish this work successfully. This material has not been published in whole or in part elsewhere and not funded by any organization or committee.
References
- Baldi P, Brunak S, Bach F, et al. 2001. Bioinformatics: The machine learning approach. MIT press.
- Murdoch TB and Detsky AS. 2013. The inevitable application of big data to health care. Jama, 309, 351-1352.
- Chang F, Dean J, Ghemawat S, et al. 2008. Bigtable: A distributed storage system for structured data. ACM Trans Comput Biol Bioinform, 26, 4.
- Raghupathi W and Raghupathi V. 2013. An overview of health analytics. J Health Med Informat, 4, 2.
- Lu Q and Cheng XH. 2012. The research of decision tree mining based on Hadoop. In 2012 9th international conference on fuzzy systems and knowledge discovery. IEEE, 798-801.
- Barga R, Fontama V, Tok WH, et al. 2015. Predictive analytics with microsoft azure machine learning. Berkely, CA: Apress.
- Kale V. 2016. Big data computing: A guide for business and technology managers, 1st edn. Chapman and Hall/CRC p. 529.
- Pavlo A, Paulson E, Rasin A, et al. 2009. A comparison of approaches to large-scale data analysis: In proceedings of the 2009 ACM SIGMOD international conference on management of data. ACM, 165-178.
- Gobel G, Andreatta S, Masser J, et al. 2001. A MeSH based intelligent search intermediary for consumer health information systems. Int J Med Inform, 64, 241-251.
- Javed Z and Afzal H. 2014. Biomedical text mining for concept identification from traditional medicine literature. In 2014 international conference on open source systems & technologies, IEEE, 206-211.
- Archenaa J and Anita EM. 2015. A survey of big data analytics in healthcare and government. Procedia Comput Sci, 50, 408-413.
- Bajcsy P, Han J, Liu L, et al. 2005. Survey of biodata analysis from a data mining perspective. In data mining in bioinformatics. Springer, 9-39.
- Chui CK, Kao B, Hung E, et al. 2007. Mining frequent item sets from uncertain data. In Pacific-Asia conference on knowledge discovery and data mining. Springer, 47-58.
- Veloso A, Meira W, Gonçalves M, et al. 2007. Multi-label lazy associative classification. In European conference on principles of data mining and knowledge discovery. Springer, 602-605.
- Yuan X, Buckles BP, Yuan Z, et al. 2002. Mining negative association rules. In proceedings ISCC 2002 seventh international symposium on computers and communications. IEEE, 623-628.
- Aggarwal CC, Li Y, Wang J, et al. 2009. Frequent pattern mining with uncertain data. In proceedings of the 15th ACM SIGKDD international conference on knowledge discovery and data mining. ACM, 29-38.
- Humbetov S. 2012. Data-intensive computing with map-reduce and hadoop. In 2012 6th international conference on Application of Information and Communication Technologies (AICT). IEEE, 1-5.
- Dean J and Ghemawat S. 2008. Mapreduce: Simplified data processing on large clusters. Commun ACM, 51, 107-113.
- Silva C and Ribeiro B. 2003. The importance of stop word removal on recall values in text categorization. In proceedings of the international joint conference on neural networks, IEEE, 3, 1661- 1666.
- Bhatia N and Jaiswal A. 2015. Trends in extractive and abstractive techniques in text summarization. Int J Comput Appl, 117.
- Juckett D. 2012. A method for determining the number of documents needed for a gold standard corpus. J Biomed Inform, 45, 460-470.
- Froud H, Benslimane R, Lachkar A, et al. 2010. Stemming and similarity measures for Arabic documents clustering. In 2010 5th international symposium on I/V communications and mobile network. IEEE, 1-4.
- Vijayarani S, Ilamathi MJ, Nithya M, et al. 2015. Preprocessing techniques for text mining-an overview. Int J Comput Sci & Nano Commun, 5, 7-16.
- Alam MM and Ismail MA. 2017. RTRS: A recommender system for academic researchers. Scientometrics, 113, 1325-1348.
- Kang N, Van Mulligen EM, Kors JA, et al. 2011. Comparing and combiningchunkers of biomedicaltext. J Biomed Inform, 44, 354-360.
- Chandrashekar G and Sahin F. 2014. A survey on feature selection methods. J Comput Eng, 40, 16-28.
- Barrios F, López F, Argerich L, et al. 2016. Variations of the similarity function of textrank for automated summarization. arXiv, 1602, 03606.
- Khan A, Baharudin B, Lee LH, et al. 2010. A review of machine learning algorithms for text-documents classification. J adv Inf Technol, 1, 4-20.
- Korde V and Mahender CN. 2012. Text classification and classifiers: A survey. Int J Artif Intell Appl, 3, 85.
- Bikku T and Paturi R. 2019. A novel somatic cancer gene based biomedical document feature ranking and clustering model: Informatics in medicine unlocked, 100188.
- Fournier VP, Lin JCW, Kiran RU, et al. 2017. A survey of sequential pattern mining: Data science and pattern recognition, 1, 54-77.
- Kamavisdar P, Saluja S, Agrawal S, et al. 2013. A survey on image classification approaches and techniques. Int J Adv Res Comput Commun Eng, 2, 1005-1009.
- Ahalya G, Pandey HM. 2015. Data clustering approaches survey and analysis: In 2015 international conference on futuristic trends on computational analysis and knowledge management (ABLAZE). IEEE, 532-537.
- Cobos C, Andrade J, Constain W, et al. 2010. Web document clustering based on global-best harmony search, K-means, frequent term sets and Bayesian information criterion: In IEEE congress on evolutionary computation. IEEE, 1-8.
- Kostakis O. 2014. Classy: Fast clustering streams of call-graphs, data mining and knowledge discovery. 28,1554-1585.