Research Article - Journal of Medical Oncology and Therapeutics (2016) Volume 1, Issue 2
YM155 Suppresses Proliferation and Survival of Multiple Myeloma Cells via Proteasomal Degradation of c-Myc
Maki Asahi1, Shigeki Ito2, Motoki Takano1 and Yoji Ishida1
1Hematology and Oncology, Department of Internal Medicine, Iwate Medical University School of Medicine, Morioka, Iwate,Japan
2Department of Medical Oncology, Iwate Medical University School of Medicine, Morioka, Iwate, Japan.
- *Corresponding Author:
- Shigeki Ito, MD, PhD
Department of Medical Oncology
Iwate Medical University, School of Medicine
19-1 Uchimaru, Morioka, Iwate 020-8505, Japan.
Tel: +81-19-651-5111 (Ext.3765)
Fax: +81-19-651-5185
E-mail: shigei@iwate-med.ac.jp
Accepted date October 05, 2016
DOI: 10.35841/medical-oncology.1.2.62-71
Visit for more related articles at Journal of Medical Oncology and TherapeuticsAbstract
The overall survival of patients with multiple myeloma has significantly improved over the last decade, mainly due to the use of novel agents such as thalidomide, lenalidomide and bortezomib; however, multiple myeloma remains incurable due to its eventual relapse, necessitating urgent development of newer agents. YM155 is a novel small molecule that suppresses transactivation of survivin and induces apoptosis in various cancer cell types. However, the effects of YM155 on multiple myeloma cells have not been fully elucidated. We herein found that YM155 inhibited proliferation of and induced caspase-dependent apoptosis in myeloma cells, partially via the suppression of survivin, myeloid cell leukemia sequence-1, and X-linked inhibitor of apoptosis protein. Interestingly, YM155 suppressed c-Myc protein expression via the proteasomal degradation. Moreover, YM155 increased F-box and WD-40 domain protein 7α expression that is a component of the Skp, Cullin, F-box-like ubiquitin ligase complex, suggesting that YM155-induced c-Myc down-regulation may be due to the proteasomal degradation through ubiquitination by F-box and WD-40 domain protein 7α. These results warrant further clinical evaluation of YM155 in MM patients.
Keywords
c-myc, Survivin, Multiple myeloma, Fbxw7, YM155.
Introduction
Multiple myeloma (MM) is a B-cell malignancy characterized by the accumulation of neoplastic plasma cells that usually secrete monoclonal proteins. The overall survival of patients with MM has significantly improved over the last decade, mainly due to the use of novel agents, such as thalidomide, lenalidomide and bortezomib, as well as incorporation of high-dose chemotherapy into autologous hematopoietic stem cell transplantation [1,2]. However, MM remains incurable as it frequently relapses even after a complete response is achieved [1,2]. Therefore, development of newer agents with distinct mechanisms of action is needed to further improve the outcomes in MM patients.
Survivin is a member of the inhibitor of apoptosis protein (IAP) family, with dual functional roles in mitosis and apoptosis [3,4]. The molecular mechanisms by which survivin impacts cell viability are not fully elucidated. However, it was shown to be essential for proper chromosome segregation and cytokinesis via its role as a subunit of the chromosomal passenger complex [5]. Conversely, the role of survivin in the inhibition of apoptosis appears to be similarly complex; it was shown to be associated with multiple parallel pathways that regulate gene expression, mitochondrial function, and proteinprotein interactions. As an example to the latter, a number of survivin’s functions are attributable to its interaction with other members of the IAP family proteins; assembly of the survivin-X-linked inhibitor of apoptosis protein (XIAP) complex in vivo abolishes XIAP-associated antiapoptotic function [6]. In addition, survivin associates with the hepatitis B X-interacting protein, and this complex, but neither protein alone, binds caspase-9 to inhibit mitochondrial apoptosis [7]. As survivin is overexpressed in many human tumors and plays a role in cancer progression and drug resistance, it is regarded as a potential therapeutic target in a variety of cancers, including MM [8,9]. In addition, it has been shown that survivin knockdown by RNA interference led to growth rate inhibition of myeloma cells related to apoptosis induction and deep cell-cycle disruption and sensitized myeloma cells to conventional antimyeloma agents [10].
These results suggest that survivin might be a rational target for myeloma therapy. YM155 is a novel small molecule that suppresses survivin transactivation and induces apoptosis in various cancer cells regardless in their p53 status [11,12]. However, the effects of YM155 on MM cells remain unclear. Here, our studies showed that YM155 induces cell growth arrest and caspase-dependent apoptosis, partially via the suppression of not only survivin but also other anti-apoptotic proteins, including myeloid cell leukemia sequence-1 (Mcl-1) and XIAP, as well as c-Myc oncoprotein in MM cells.
Materials and Methods
Reagents and Antibodies
The survivin inhibitor YM155 and BET bromodomain inhibitor JQ1 were purchased from Selleck Chemicals (Houston, TX) and the proteasome inhibitor MG132 was purchased from EMD Biosciences (La Jolla, CA). Both agents were dissolved in DMSO as a stock solution and working solutions were prepared from the stock in RPMI 1640 medium. Control cells received 0.1% DMSO in complete medium (the maximum final DMSO concentration in YM155 treated cells). Antibodies against survivin, Mcl-1, XIAP, cleaved caspase-3, cleaved poly ADP-ribose polymerase (PARP), interferon regulatory factor (IRF)-4, heat shock protein (Hsp) 90 and β-actin were obtained from Cell Signaling Technology (Beverly, MA). Antibodies against B-cell lymphoma-2 (Bcl-2) and c-Myc were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against F-box and WD-40 domain protein 7α (Fbxw7α) were purchased from Allele Biotechnology (San Diego, CA).
Cell Lines
The human RPMI8226 myeloma cell line, obtained from the American Type Culture Collection (ATCC, Rockville, MD), was originally derived from the peripheral blood of a 61 year old male with MM [13]. The human U266 myeloma cell line kindly provided by Dr. Kuroda (Hiroshima University, Hiroshima, Japan) is an IgEproducing cell line derived from a 53 year male with MM [14]. The KMS-28PE, KMS-20 and KMS-34 myeloma cell lines were kindly provided by Dr. Tamura (Nippon Medical School, Tokyo, Japan). KMS-28PE and KMS-34 were derived from the pleural effusion of a 77 year female and a 60 year female with t(4;14) myeloma, respectively. KMS-20 was derived from a 65 year female with MM. These cell lines were originally established at Kawasaki Medical School, Okayama, Japan [15,16]. MM cell lines were cultured in RPMI1640 (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 100 U/ml penicillin and 100 μg/ ml streptomycin (Gibco), and 2 mM of glutamine (Gibco) in a 5% CO2 incubator at 37°C.
Cell Proliferation Assay
Cell proliferation assay was performed as described previously [12]. Briefly, 2 × 105/ml cells from each line were cultured in RPMI1640 medium supplemented with 10% FBS in the presence or absence of indicated concentrations of YM155 for 24 h and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed according to the manufacturer’s recommendations. For cell proliferation assay using JQ1, RPMI8226, KMS28-PE, and KMS-34 cells (5 × 105/ml) were cultured in complete RPMI8226 medium in the presence of 0, 0.5 and 1 μM JQ1 in 12-well tissue-culture plates (BD Labware, Franklin Lakes, NJ). After 24 h of culture, the cell numbers were determined by the trypan-blue (Gibco, Grand Island, NY) exclusion method [17].
Apoptosis Assay
Apoptosis assay was performed as described previously [12]. Briefly, RPMI8226 and KMS-28PE cells were plated at a concentration of 2 × 105/ml with or without 10 nM YM155 for 24 or 48 h. In addition, to confirm dosedependent cell death, KMS-28PE cells were incubated with 0 nM, 10 nM or 100 nM YM155 for 24 h. Two hundred thousand cells were stained with annexin V and propidium iodide (PI) according to the manufacturer’s instructions (BD Biosciences, San Diego, CA). The percentage of annexin V-positive cells was analyzed by the FACS calibur (Becton Dickinson, LA).
cDNA Synthesis And Real-Time Reverse Transcription- Polymerase Chain Reaction
cDNA synthesis and real-time reverse transcription (RT)- polymerase chain reaction (PCR) were performed as described previously [12]. RPMI8226, KMS-28PE and KMS-34 cells were cultured in the presence or absence of YM155 for 6 h. One microgram total RNA isolated using RNeasy Mini Kit (Qiagen Sciences, MD) from each condition was reverse transcribed using first-strand cDNA synthesis kit (Qiagen), which was then assayed by realtime reverse transcription (RT)-polymerase chain reaction (PCR) (Light-Cycler, Roche Applied Science, Indianapolis, IN) according to the technical brochure of the company. The primers used for human c-myc were as follows: forward, 5′-CTTCTCTCCGTCCTCGGATTCT-3′; and reverse, 5′-GAAGGTGATCCAGACTCTGACCTT-3′. The primers for GAPDH as internal control were purchased from Nihon Gene Research Laboratories Inc. (Sendai, Japan). SYBR green (Roche Diagnostics, Indianapolis, IN) was used as the PCR probe and the ratio of c-myc to GAPDH was calculated from the sample studies.
Immunoblotting
Immunoblotting was performed as described previously [18]. After treatment with YM155, RPMI8226, KMS-28PE and KMS-34 cells were washed twice with cold phosphatebuffered saline (PBS), and protein lysates were prepared by incubating pellets in ice-cold lysis buffer (0.5% NP-40, 10 mM Tris base, 200 mM NaCl, 10% glycerol, 5 mM NaF, 0.5 mM sodium orthovanadate, pH 7.4) supplemented with 1% protease inhibitor cocktail (EMD Biosciences, San Diego, CA) for 30 min at 4°C. The supernatants were cleared by centrifugation at 4066 rpm for 10 min and eluted by incubation with SDS sample buffer (Pierce, Rockford, IL) for 5 min at 95°C. The samples were loaded on 7%-15% Tris-glycine gels (Bio-Rad Laboratories, Hercules, CA) and transferred to polyvinylidene difluoride membranes (GE Healthcare, Buckinghamshire, UK). Immunoblotting was then performed according to the antibody manufacturers’ instructions and visualization was achieved with HRP-conjugated secondary antibodies and the enhanced chemiluminescent detection system (ECL plus; GE Healthcare). In certain experiments, membranes were incubated in a stripping buffer (Pierce, Rockford, IL) according to the manufacturer’s instructions, before reprobing with new primary antibodies.
Proteasome Inhibition Assay
Proteasome inhibition assay was performed as described previously [19]. RPMI8226 and KMS-28PE cells were treated in the absence or presence of either YM155 (50 nM), MG-132 (10 μM) or both for 6 h. Immunoblot analyses with anti-c-Myc, anti-IRF4 and anti-β-actin antibodies were performed as described above.
Statistical Analysis
JMP® 11 (SAS Institute, Cary, NC) was used for statistical analysis. The data obtained were expressed as means ± standard deviation (SD). Differences between test groups were analyzed by the Wilcoxon test. P-values <0.05 were considered statistically significant.
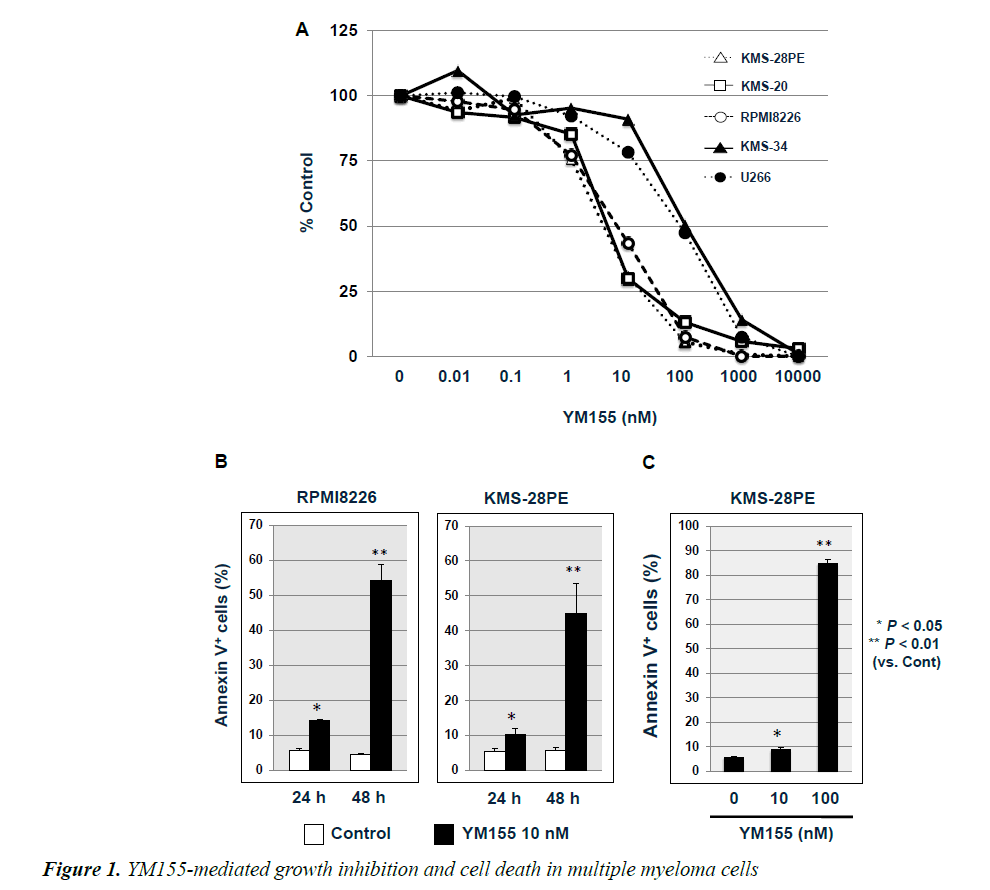
Figure 1. YM155-mediated growth inhibition and cell death in multiple myeloma cells.
(A) KMS-28PE, KMS-20, KMS-34, RPMI8226, and U266 cells were cultured in a 96-well tissue culture plates in the absence or presence of YM155 at the indicated concentrations for 24 h. Cell viability was measured using the MTT assay. Data are expressed as the mean percentage of the control cells, representing the average of triplicate samples. Results are representative of three independent experiments. (B) RPMI8226 and KMS-28PE cells were cultured in a 6-well tissue culture plates in the absence or presence of YM155 (10 nM) for 24 h or 48 h, respectively, without medium exchange. The percentage of cell death was measured using flow cytometric analysis of annexin-V/propidium iodide (PI) staining. Data represent the average of triplicate samples, and error bars represent one standard deviation (SD) from the mean for triplicate wells. Results are representative of two independent experiments. (C) KMS-28PE cells were cultured in a 6-well tissue culture plates in the absence or presence of YM155 (10 nM or 100 nM) for 24 h. Data represent the average of triplicate samples, and error bars represent one SD from the mean for triplicate wells. Results are representative of two independent experiments (YM155 versus control, *P<0.05, **P<0.01).
Results
YM155 Suppresses Cell Proliferation and Survival in MM Cell Lines
We examined whether YM155 suppressed the proliferation of MM cell lines. As shown in Figure 1A, YM155 treatment inhibited the proliferation of five tested MM cell lines in a dose-dependent manner. However, the sensitivity was different among these cell lines; RPMI8226, KMS- 28PE and KMS-20 cells were more sensitive to YM155 than KMS-34 and U266 cells (IC50=10 and 100 nM, respectively). Because we wanted to know more clearly the effects of YM155 on MM cells, we used two YM155- sensitive MM cell lines (RPMI8226 and KMS-28PE) in the cell death assay. We next examined whether YM155 induced cell death in RPMI8226 and KMS-28PE cell lines, by measuring the percentage of cell death using flow cytometric analysis with annexin V and PI staining after cells were cultured in the presence or absence of 10 nM YM155 for 24 h or 48 h. As shown in Figure 1B, the percentage of annexin V (+) cells was higher in cultures treated with YM155 than those not treated with the survivin inhibitor. In addition, the significantly higher percentage of annexin V (+) cells in KMS-28PE cultures incubated with 100 nM YM155 compared to those exposed to 10 nM YM155 (P<0.01) over 24 h indicated that YM155- mediated cell death was occurring in a dose-dependent manner (Figure 1C). These results suggested that YM155 suppressed proliferation and induced cell death in MM cell lines.
YM155 Suppresses Survivin, Mcl-1 and XIAP Expression in RPMI8226 and KMS-28PE Cells
To better elucidate the mechanisms by which YM155 suppressed proliferation and induced cell death in MM lines, we examined the expression of anti-apoptotic proteins in RPMI8226 and KMS-28PE cells. As shown in Figure 2, the protein levels of survivin, Mcl-1 and XIAP were suppressed 24 h after treatment with YM155 in both cell lines, whereas there was no change in the expression of Bcl-2. In addition, YM155 activated caspase-3 and PARP in both cell lines. These results suggested that YM155 induced caspase-dependent apoptosis in RPMI8226 and KMS-28PE MM cell lines.
YM155 Suppresses C-Myc and IRF-4 Expression in Myeloma Cells
We next examined the effects of YM155 on the expression of c-Myc and IRF-4, both of which play a crucial role in pathogenesis of MM [20]. Interestingly, both c-Myc and IRF-4 protein levels were suppressed following 24 h treatment of RPMI8226 and KMS-28PE cells with YM155. In contrast, IRF-4 level was not suppressed in KMS-34 cells although c-Myc level was suppressed (Figure 3A). As c-Myc and IRF-4 were shown to form a positive feedback loop in MM cells, we next determined which transcription factor was first suppressed by YM155.
As shown in Figure 3B, YM155 led to reductions in c-Myc protein levels at 6 h after treatment, whereas IRF-4 protein reductions were evident only at least after 12 h in RPMI8226 but not even after 18 h in KMS-28PE and KMS-34 cells, suggesting that YM155 suppressed c-Myc expression followed by IRF-4 in RPMI8226 and KMS-28PE cells.
Figure 2. YM155 suppresses survivin, Mcl-1, and XIAP expression and activates caspase-3 in multiple myeloma cells. RPMI8226 and KMS-28PE cells were treated with YM155 (10 nM or 100 nM) for 24 h. Immunoblotting was performed with specific antibodies against survivin, cleaved caspase-3, cleaved PARP, XIAP, Mcl-1, Bcl-2 and ß-actin. The immunoblot shown is representative of two independent experiments.
Figure 3. YM155 suppresses c-Myc and IRF-4 expression in multiple myeloma cells. (A) RPMI8226, KMS-28PE and, KMS-34 cells were treated with YM155 (10 nM or 100 nM) for 24 h. (B) RPMI8226, KMS-28PE, and KMS-34 cells were treated with YM155 (10 nM or 100 nM) for 0 h, 6 h, 12 h or 18 h. Immunoblotting was performed with specific antibodies against c-Myc, IRF- 4, and ß-actin. The immunoblot shown is representative of two independent experiments.
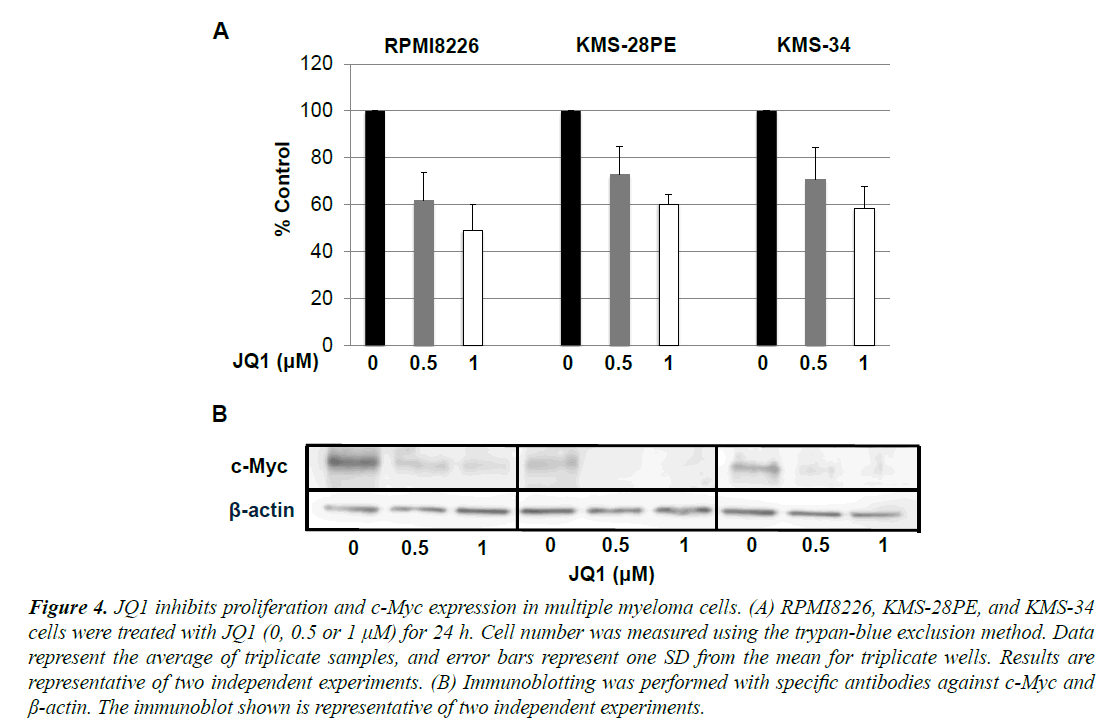
We next examined if the proliferation of myeloma cell lines depends on c-Myc. As shown in Figure 4A, the BET bromodomain inhibitor JQ-1 which has recently shown to down-regulate c-myc transcription suppressed proliferation [21], but not fully, in RPMI-8226, KMS-28PE and KMS-34 cells in a dose-dependent manner.
Western blot analysis showed that JQ-1 inhibited c-Myc protein expression in the same manner (Figure 4B). These results suggested that the proliferation in these myeloma cells depends at least in part on c-Myc.
MG132 Blocks YM155-Induced Down-Regulation of C-Myc in RPMI8226 and KMS-28PE Cells
Figure 4. JQ1 inhibits proliferation and c-Myc expression in multiple myeloma cells. (A) RPMI8226, KMS-28PE, and KMS-34 cells were treated with JQ1 (0, 0.5 or 1 µM) for 24 h. Cell number was measured using the trypan-blue exclusion method. Data represent the average of triplicate samples, and error bars represent one SD from the mean for triplicate wells. Results are representative of two independent experiments. (B) Immunoblotting was performed with specific antibodies against c-Myc and ß-actin. The immunoblot shown is representative of two independent experiments.
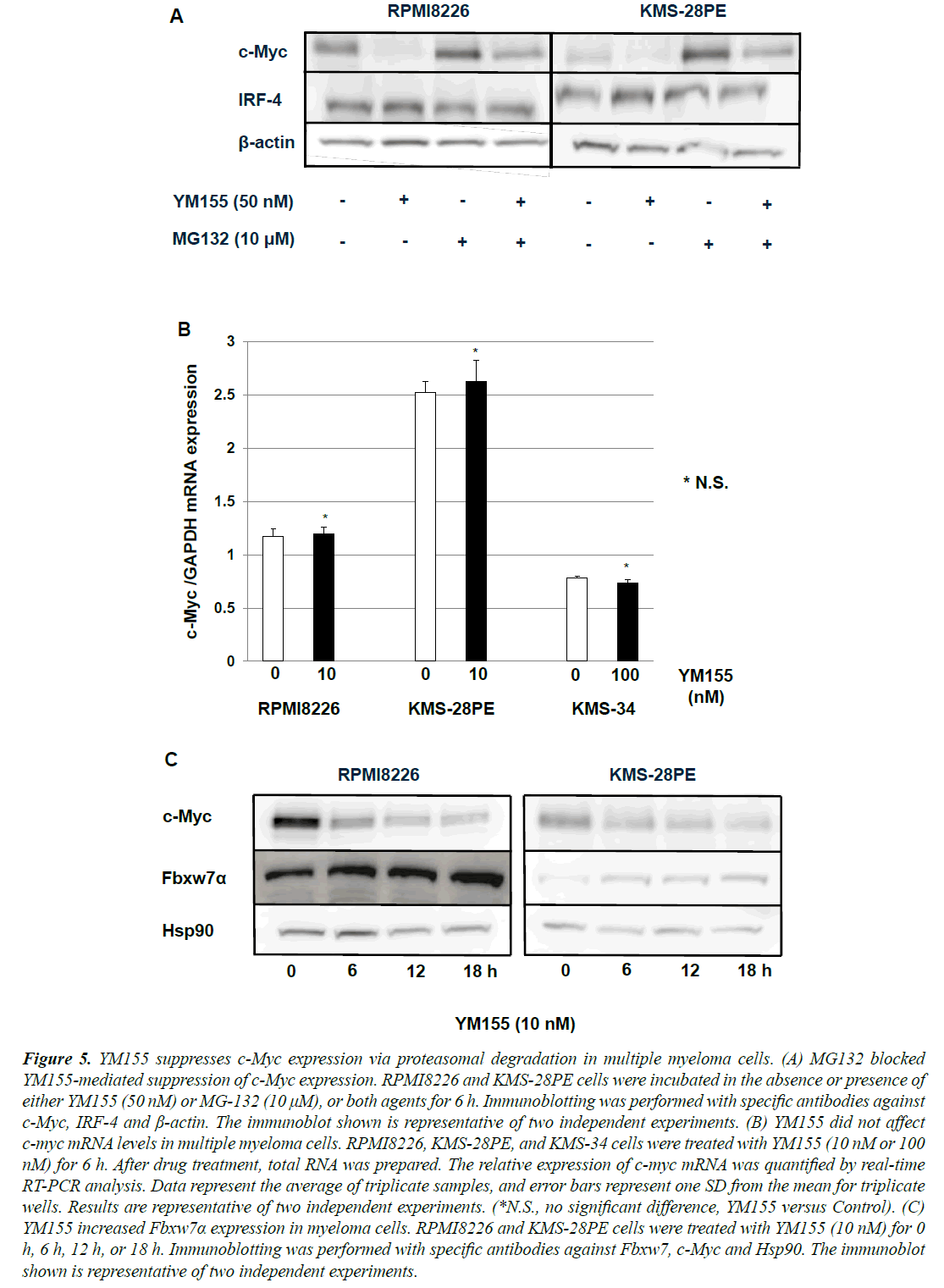
Figure 5. YM155 suppresses c-Myc expression via proteasomal degradation in multiple myeloma cells. (A) MG132 blocked YM155-mediated suppression of c-Myc expression. RPMI8226 and KMS-28PE cells were incubated in the absence or presence of either YM155 (50 nM) or MG-132 (10 µM), or both agents for 6 h. Immunoblotting was performed with specific antibodies against c-Myc, IRF-4 and ß-actin. The immunoblot shown is representative of two independent experiments. (B) YM155 did not affect c-myc mRNA levels in multiple myeloma cells. RPMI8226, KMS-28PE, and KMS-34 cells were treated with YM155 (10 nM or 100 nM) for 6 h. After drug treatment, total RNA was prepared. The relative expression of c-myc mRNA was quantified by real-time RT-PCR analysis. Data represent the average of triplicate samples, and error bars represent one SD from the mean for triplicate wells. Results are representative of two independent experiments. (*N.S., no significant difference, YM155 versus Control). (C) YM155 increased Fbxw7a expression in myeloma cells. RPMI8226 and KMS-28PE cells were treated with YM155 (10 nM) for 0 h, 6 h, 12 h, or 18 h. Immunoblotting was performed with specific antibodies against Fbxw7, c-Myc and Hsp90. The immunoblot shown is representative of two independent experiments.
Cells employ numerous mechanisms, including proteolysis, to control the expression and degradation of c-Myc as it is an important regulator of genes required for cell growth and proliferation. To elucidate the mechanism of YM155- induced suppression of c-Myc protein expression, we first performed a proteasome inhibition assay. As shown in Figure 5A, treatment with a proteasome inhibitor MG132 blocked YM155-mediated decreases in c-Myc expression in RPMI8226 and KMS-28PE cells, suggesting that the degradation of c-Myc might be occurring via the ubiquitinproteasome pathway.
We next examined whether YM155 repressed the transcription of c-myc oncogene. As shown in Figure 5B, with quantitative RT-PCR analysis, c-myc mRNA levels were not decreased at 6 h after treatment with 10 nM YM155 in RPMI8226 and KMS-28PE cells and with 100 nM YM155 in KMS-34 cells. These results indicated that YM155 might reduce c-Myc expression mainly via the ubiquitin-proteasome pathway.
We next investigated how YM155 promotes c-Myc degradation in myeloma cells. Because c-Myc is one of the targets of Fbxw7α that is a component of the SCF-like ubiquitin ligase complex [22], we examined if YM155 treatment up-regulates Fbxw7α expression. As shown in Figure 5C, YM155 increased Fbxw7α expression in RPMI8226 and KMS-28PE cells, suggesting that YM155 might promote the proteasomal degradation of c-Myc at least in part via ubiquitination by Fbxw7α.
Discussion
Survivin is frequently overexpressed in a variety of cancer types [23]. Specifically in MM, its overexpression is detectable in 43% of patient biopsy samples and is associated with not only adverse chromosome aberrations such as 17p deletion and 1q21 gain, but also adverse prognosis such as worse event-free and overall survival rates. Wagner et al. showed that survivin expression was associated with cell proliferation, adverse prognostic markers, and inferior event-free and overall survival rates in their cohort [24]. In addition, Romagnoli et al. reported that survivin was a direct contributor to the malignant progression of MM and that higher survivin expression correlated with poorer survival in MM patients [25].
Overall, these findings suggest survivin as a potential target in patients with high-risk MM. In this study, we showed that survivin inhibitor YM155 suppressed the proliferation of MM cells and induced apoptosis in a dose- and timedependent manner. In addition, YM155 down-regulated survivin, XIAP and Mcl-1 expression and activated caspase-3 and PARP, are indicating YM155-induced caspase-dependent apoptosis in these cells. These findings were compatible with recent report that YM155 induced cell growth arrest and apoptosis via down-regulating Mcl-1 expression in myeloma cells [24]. Mcl-1 is a Bcl- 2 family member and plays critical roles in promoting MM cell survival via its anti-apoptotic functions [26,27]. The expression of Mcl-1 in MM cells is tightly regulated by survival signaling triggered by cytokines and growth factors in the bone marrow microenvironment [26,28,29]. Feng et al. also demonstrated YM155-mediated survivin and Mcl-1 downregulation in human leukemia cells [30].
In that study, YM155 induced apoptosis concomitant with the activation of caspase-8 and caspase-3. The bone marrow microenvironment contributes to resistance against various therapies in MM. This is hypothesized to occur through the interaction of MM cells with accessory cells of the microenvironment, leading to the induction of pleiotropic anti-apoptotic mechanisms. A recent report demonstrated that adhesion to accessory cells upregulated survivin within MM cells. They also determined that bone marrow stroma cells from patients with MM and healthy individuals as well as vascular endothelial cells significantly inhibited the cytotoxic T-lymphocyte (CTL)– mediated lysis of MM cells. In that study, the authors showed that repression of survivin with YM155 improved CTL-mediated lysis of MM cells in vitro and in vivo in the presence of accessory cells [31]. These results revealed the cell adhesion-mediated induction of apoptosis resistance as a novel immune escape mechanism and provided a rationale to improve the efficacy of cellular therapies by pharmacologic modulation of cell adhesion-mediated immune resistance. Overall, these studies suggested that YM155 might manifest its antitumor activity through not only survivin but also Mcl-1 inhibition in MM cells.
As an oncogene c-myc has pivotal roles in cell growth, proliferation, tumorigenesis and metabolism [32]. The importance of c-myc activation in MM has been shown through the use of the Vk*MYC transgenic mouse model, where activation of c-myc occurs through activationinduced cytidine deaminase (AID)-dependent somatic hypermutation during B-cell development, which results in myeloma onset in these mice [33]. In addition, MM cells were shown to depend on c-Myc for survival, which was shown by induction of cell death via ablation of c-myc by small hairpin RNAs or small molecule inhibitors [34]. In our study, c-Myc inhibitor JQ1 suppressed cell proliferation and c-Myc protein expression. Walker et al. recently reported that MM patients with c-myc translocations had decreased progression-free and overall survival rates [35].
Overall, these findings indicate c-myc as a promising therapeutic target in MM. Unfortunately, despite evergrowing understanding of the biology of c-myc; targeted clinical therapeutic strategies are not available for tumors with increased c-myc expression. Our study clearly illustrated YM155-mediated suppression of c-Myc in MM cells. In our studies investigating the underlying mechanism, we observed that the expression of both c-Myc and IRF-4 were inhibited in RPMI8226 and KMS- 28PE cells. A recent study demonstrated a positive autoregulatory loop between IRF-4 and c-Myc during normal B-cell activation as well as in MM [20]. In our study, change in c-Myc preceded that of IRF-4, indicating c-Myc inhibition prior to IRF-4 by YM155. Our results also showed that the recovery of YM155-mediated suppression of c-Myc protein levels by MG132, whereas c-myc mRNA levels were not inhibited after 6 h treatment with YM155 in MM cells.
Taken together, these results suggest that YM155 might suppress c-Myc expression at least in part via the proteasomal degradation pathway in MM cells. Several different E3 ligases have been implicated in regulating c-Myc protein stability through the ubiquitin-proteasome system. Recent report demonstrated that Fbxw7 functions as a component of SCF-like ubiquitin ligases that catalyzes the ubiquitination of c-Myc [22]. In our study, we found that YM155 increased Fbxw7 expression in RPMI8226 and KMS-28 cells. These results suggest that YM155- induced c-Myc down-regulation may be due to the proteasomal degradation through ubiquitination by Fbxw7 in myeloma cells. However, the precise mechanism of YM155-mediated Fbxw7 induction remains unclear and warrants further investigation in future studies.
Because the single agent activity of YM155 appears to be modest in the clinical trial, appropriate combination with novel anti-myeloma agent may be required in the clinical setting. Bortezomib is one of the key drugs for myeloma treatment. Recent report demonstrated that the combination of YM155 with bortezomib showed moderate antagonistic effects in myeloma cell lines. In contrast, co-treatment of myeloma cells with YM155 and the antagonist of Bcl- 2 proteins, ABT-263 led to a marked synergistic growth inhibition [24]. These results support the early evaluation of YM155 in combination with inhibitors of other Bcl-2 family members in clinical trials.
Conclusion
The survivin inhibitor YM155 induces caspase-dependent apoptosis at least in part via inhibiting survivin, Mcl-1 and XIAP expressions in MM cells. In addition, YM155 suppresses c-Myc oncoprotein, which plays a critical role in MM pathogenesis, via the proteasomal degradation pathway. Our data indicate that clinical evaluation of YM155 may be warranted in patients with MM.
Acknowledgement
We thank to Ms. Shizune Hirayama and Ms. Hiroko Chiba for technical assistance. M.A. and M.T. and S.I. performed experiments. M.A. and S.I. and Y.I. designed research, wrote the paper and discussed the manuscript.
Conflict of Interest
The authors declare no competing financial interests.
References
- Kumar SK, Rajkumar SV, Dispenzieri A, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood 2008;111:2516-2520.
- Kumar SK, Dispezieri A, Lacy MQ, et al. Continued improvement in survival in multiple myeloma: Changes in early mortality and outcomes in older patients. Leukemia 2014;28:1122-1128.
- Ambrosini R, Adida C, ALtieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997;3:917-921.
- Colnaghi R, Connel CM, Barrett RM, et al. Separating the anti-apoptotic and mitotic roles of survivin. J BiolChem 2006;281:33450-33456.
- Jeyaprakash AA, Klein UR, Lindner D, et al. Structure of a survivin-borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 2007;131:271-285.
- Dohi T, Xia F, Altieri DC. Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection.Mol Cell 2007; 27: 17-28.
- Marusawa H, Matsuzawa S, Welsh K, et al. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J 2003; 22: 2729-2740.
- Tsubaki M, Takeda T, Ogawa N, et al. Overexpression of survivin via activation of ERK1/2, Akt and NF-kB plays a central role in vincristine resistance in multiple myeloma cells. Leuk Res 2015;39:445-452.
- Hideshima T, Catley L, Raje N, et al. Inhibition of Akt induces signify downergulation of survivin and cytotoxicity in human multiple myeloma cells. Br J Haematol 2007;138:783-791.
- Romagnoli M, Trichet V, David C, et al. Significant impact of survivin on myeloma cell growth. Leukemia 2007;21:1070-1078.
- Nakahara T, Kita K, Yamanaka M, et al. YM155, novel small-molecule survivin suppressant, induces regression of established human-hormone refractory prostate tumor xenografts. Cancer Res 2007;67:8014-8021.
- Sasaki R, Ito S, Asahi M, et al. YM155 suppresses cell proliferation and induces cell death in human adult T-cell leukemia/lymphoma cells. Leuk Res 2015;39:1473-1479.
- Matsuoka Y, Moore GE, Yagi Y, et al. Production of free light chains of immunoglobulin by a hematopoietic cell line derived from a patient with multiple myeloma. ProcSocExp Med 1967;125:1246-1250.
- Nilsson K, Bennich H, Johansson SG, et al. Established immune producing myeloma (IgE) and lymphoblastoid (IgG) cell lines from an IgE myeloma patient. ClinExpImmunol 1970;7:477-489.
- Otsuki T, Yata K, Sakaguchi H, et al. Interleukin 10 abolishes the growth inhibitory effects of all-trans retinoic acid on human myeloma cells. Br J Haematol 2002;116:787-795.
- Miura Y, Tsujioka T, Nishimura Y, et al. TRAIL expression up-regulated by interferon-gamma via phosphorylation of STAT1 induces myeloma cell death. Anticancer Res 2006;26:4115-4124.
- Esumi H, Izushi K, Kato K, et al. Hypoxia and nitric oxide treatment confer tolerance to glucose starvation in a 5?-AMP-activated protein kinase-dependent manner. J BiolChem 2002;277:32791-32798.
- Ito S, Mantel C, Han MK, et al. Mad2 is required for optimal hematopoiesis: Mad2 associates with c-Kit in MO7e cells. Blood 2007;109:1923-1930.
- Ito S, Oyake T, Murai K, et al.Deguelin suppresses cell proliferation via the inhibition of survivin expression and STAT3 phosphorylation in HTLV-1-transformed T cells. Leuk Res 2010;34:352-357.
- Shaffer AL, Emre NC, Lamy L, et al. IRF4 addiction in multiple myeloma. Nature 2008;454:226-231.
- Delmore JE, Issa G, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904-917.
- Yada M, Hatakeyama S, Kamura T, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbx7. EMBO J 2004;23:2116-2125.
- Velculescu VE. Essay: Amersham Pharmacia Biotech & Science prize. Tantalizing transcripts?SAGE and its use in global gene expression analysis. Science 1999;286:1491-1492.
- Wagner V, Hose D, Seckinger A, et al. Preclinical efficacy of sepantronium bromide (YM155) in multiple myeloma is conferred by down regulation of Mcl-1. Oncotarget 2014;5:10237-10250.
- Romagnoli M, Séveno C, Wuillème-Toumi S, et al. The imbalance between survivin and Bim mediates tumor growth and correlates with poor survival in patients with multiple myeloma. Br J Haematol 2009;145:180-189.
- Le Gouill S, Podar K, Harousseau JL, et al. Mcl-1 regulation and its role in multiple myeloma. Cell Cycle 2004;3:1259-1262.
- Perciavallé RM, Opferman JT. Delving deeper: MCL-1?s contributions to normal and cancer biology. Trends Cell Biol 2013;23:22-29.
- Thomas LW, Lam C, Edwards SW. Mcl-1: The molecular regulation of protein function. FEBS Lett 2010;584:2981-2989.
- Huston A, Roodman GD. Role of the microenvironment in multiple myeloma bone disease. Future Oncol 2006;2:371-378.
- Feng W, Yoshida A, Ueda T. YM155 induces caspase-8 dependent apoptosis through downergukation of survivin and Mcl-1 in human leukemia cells. BiochemBiophys Res Commun 2013;435:52-57.
- deHaart SJ, van de Donk KW, Minnema MC, et al. Accessory cells of the microenvironment protect multiple myeloma from T-cell cytotoxicity through cell adhesion-mediated immune resistance. Clin Cancer Res 2013;19:5591-5601.
- Dang CV. MYC on the path to cancer. Cell 2012;149:22-35.
- Chesi M, Robbiani DF, Sebag M, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 2008;13:167-180.
- Holien T, Vatsveen TK, Hella H, et al. Addiction to c-MYC in multiple myeloma. Blood 2012;120:2450-2453.
- Walker BA, Wardell CP, Brioli A, et al. Translocations at 8q24 juxtapose MYC with genes that harbor supermenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer Journal 2014;4:e191.