Research Article - Biomedical Research (2016) Health Science and Bio Convergence Technology: Edition-I
Total RNA yield and its correlation with GAPDH expression in liver and kidneys of rats treated with gold nanoparticles
Haseeb A. Khan1*, Salman H. Alrokayan1, Abdullah S. Alhomida1, Isra Khan21Department of Biochemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
2Rohilkhand Medical College and Hospital, Bareilly, India
- *Corresponding Author:
- Haseeb A. Khan
Department of Biochemistry
College of Science, King Saud University
Saudi Arabia
Accepted date: December 06, 2016
Abstract
Gold Nanoparticles (GNPs) are gaining more attention in the field of nanomedicine, particularly in targeted drug delivery and diagnostic probes. More recently, GNPs have been applied in the emerging area of RNA nanotechnology. We studied the effect of 10 nm and 50 nm GNPs on total RNA yield from liver and kidneys of rats. The rats were intraperitoneally injected with 10 nm or 50 nm GNPs for 1 or 5 days and the samples of livers and kidneys were collected 24 h after the last injection. Small portions of tissues were immediately submerged in RNAlater RNA stabilization reagent for in situ stabilization of cellular RNA. Total RNA was extracted using RNAeasy kit and the yield and purity of RNA were measured using a nanodrop spectrophotometer. The total RNA yield was several folds higher from liver as compared to kidneys of rats. Treatment of rats with 10 nm and 50 nm GNPs did not affect the RNA yield from liver whereas the 50 nm GNPs significantly reduced the RNA yield from the kidneys of rats. There was a significant decrease in GAPDH expression in liver and kidney of rats treated with 50 nm GNPs, at day 1 post-dosing.
Keywords
RNA yield, GAPDH expression, Liver, Kidney, Gold nanoparticles, Rats.
Introduction
In recent years, the production of engineered nanoparticles has progressively increased due to their applications in various fields such as medicine, electronics, cosmetics, bioremediation, coatings, paints, food industry and water treatment [1-8]. However, massive use of nanoparticles without knowing their toxic effects might pose potential hazardous impact on human health and environment. More recently, RNA nanoparticles are gaining interests due to their potential applications in nanomedicine [9,10]. Notwithstanding much technological advancement, our knowhow about the toxicological effects of nanoparticles is quite narrow and so far there are no specific guidelines about the use of engineered nanoparticles. Being the two sides of the same coin, nanomedicine and nanotoxicology should be prudently applied in order to maintain the worth of this coin [11].
The unique properties of Gold Nanoparticles (GNPs) on the aspects of biocompatibility, resistance to oxidation, high surface reactivity and flexibility in functionalization render them as promising therapeutic and diagnostic tools in nanomedicine [12,13]. GNPs modified by branched polyethyleneimine have been used as efficient and safe intracellular delivery carriers for siRNA [14]. GNPs-assisted photo-controlled intracellular RNA delivery has been reported to be a promising strategy with high target specificity [15]. Recently, magnetic GNPs have been applied for delivery of siRNA for efficient silencing of oncogenes [16]. Jackson et al. [17] have developed DNA-functionalized GNPs for multiplexed detection of mRNA expression in live breast cancer cells using flow cytometry and fluorescence microscopy. Yang et al. [18] have reported a sensitive and specific detection method for quantification of microRNA using GNP probe. Exposure of GNPs has been shown to alter the expression of mRNA of various genes both in-vivo and invitro [19-25]. In this investigation, we studied the effect of GNPs on total RNA yield from liver and kidneys of rats.
Materials and Methods
Animals and treatment groups
The study was conducted on adult male Wistar rats (body weight 230 ± 20 g), obtained from the Laboratory Animal Centre, College of Pharmacy, King Saud University, Riyadh. The animals were maintained in a humidity- and temperaturecontrolled room with 12 h light/dark cycles and with free access to standard laboratory food and tap water. The animals were randomly divided into 5 groups of 5 animals each. Group 1 served as control and received vehicle only. Two groups were treated with GNPs (10 nm diameter) for 1 day (Group 2) or daily for 5 days (Group 3). The remaining two groups received GNPs (50 nm diameter) for 1 day (Group 4) or daily for 5 days (Group 5).
Dosing of gold nanoparticles
Commercially available GNPs of 10 nm diameter (MKNAu- 010 of concentration 0.01% Au) and 50 nm (MKN-Au-050 of concentration 0.01% Au) were purchased from MK Impex Corp., Ontario, Canada. Doses of 50 μl of 10 nm and 50 nm GNPs in aqueous solution were administered to animals via intraperitoneal injection daily for 1 or 5 days. This dose regimen was approximately equivalent to 5 μg/animal of 10 nm GNP (number of particles, 2.85 × 1011) or 50 nm GNP (number of particles, 2.25 × 109). All experiments were conducted in accordance with guidelines approved by our Institutional Animal Care and Use Committee.
Sampling and storage
The rats were sacrificed 24 h after the last injection of GNPs and specimens of liver and kidneys were isolated. Immediately after harvesting, small portions of tissues (approx. 30 mg) were immediately submerged in RNAlater RNA Stabilization Reagent (Qiagen), which rapidly permeates the tissues to stabilize and protect cellular RNA in situ. Tissues protected in RNAlater were stored at 4°C until RNA extraction. The RNAlater technology allows large numbers of samples to be easily processed and replaces inconvenient, dangerous, and equipment-intensive methods, such as snap-freezing of samples in liquid nitrogen, storage at-80°C, cutting and weighing on dry ice, or immediate processing of harvested samples.
Purification of total RNA from liver and kidney
We used RNAeasy kit (Qiagen) for purification of total RNA from animal tissues. The RNeasy procedure represents a microspin column-based technology for RNA purification, using the selective binding properties of a silica-based membrane and successive centrifugations. The RNAlaterstabilized tissue (approx. 20 mg) was disrupted in 600 μl Buffer RLT (contains guanidine thiocyanate and β- mercaptoethanol) and homogenized using Ultraturax homogenizer. The homogenate was centrifuged at 10,000 rpm for 3 min and the supernatant was carefully removed and transferred to a new microcentrifuge tube. An equal volume of 70% ethanol was added to the clear supernatant and mixed immediately by pipetting. Ethanol helps in creating conditions that promote selective binding of RNA to the RNeasy membrane. Up to 700 μl of the above solution was transferred to an RNeasy spin column placed in a 2 ml collection tube and centrifuged at 10,000 rpm for 15 s. After discarding the flowthrough, 700 μl of Buffer RW1 were added to the RNeasy spin column and centrifuged at 10,000 rpm for 15 s to wash the spin column membrane. After discarding the flow-through, 500 μl of Buffer RPE were added to the RNeasy spin column and centrifuged at 10,000 rpm for 15 s to wash the spin column membrane. RPE washing was repeated with the same volume of buffer but increased centrifugation time (2 min). The RNeasy spin column was removed and placed in a new 1.5 ml collection tube. After adding 30 μl RNase-free water to the spin column membrane, the tube was centrifuged at 10,000 rpm for 1 min to elute the RNA.
Determination of RNA yield and purity
The RNA yield and purity were determined spectrophotometrically using a Nanodrop spectrophotometer (Thermo Fisher). RNA is known to have a maximum absorption (λmax) at 260 nm. An absorbance at 260 nm (A260) reading of 1.0 is equivalent to about 40 μg/ml of RNA, was used to determine the RNA concentration in the solution. To assess the purity of RNA preparation, the ratio of the absorbance at 260 and 280 nm was used. Pure RNA has an A260/A280 of around 2.0.
Gene expression analysis
We used one of the house-keeping genes, Glyceraldehyde-3- Phosphate Dehydrogenase (GAPDH), to determine the integrity of extracted RNA. The GAPDH gene expression was analysed by real-time PCR in a 20 μl reaction mixture containing 0.25 μM of forward and reverse primers, 2 μl of RNA template and 12.5 μl SybrGreen real-time PCR MasterMix (Applied Biosystems, USA). The reaction mixture was incubated at 50°C for 30 min and then at 95°C for 10 min, for reverse transcription and polymerase enzyme activation, respectively. The subsequent cycling conditions were as follows: 45 cycles of 95°C for 15 s, 60°C for 20 s, and 72°C for 60 s. The GAPDH primers sequences were: forward primer, 5-GTA TTG GGC GCC TGG TCA CC-3 and reverse primer, 5-CGC TCC TGG AAG ATG GTG ATG G-3.
Statistics
The data were analysed by one-way Analysis of Variance (ANOVA) followed by Dunnett’s multiple comparison test using SPSS statistical package. Pearson’s test was used for correlation study. P values<0.05 were considered as statistically significant.
Results
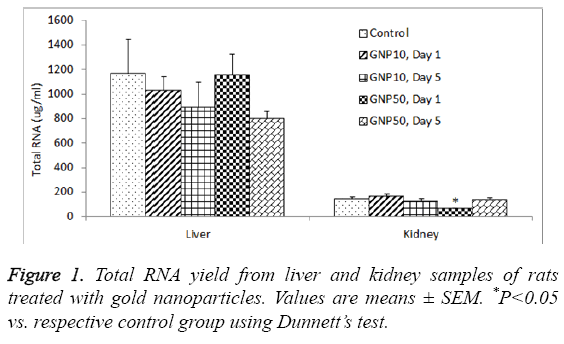
The total RNA yield from the control rat liver was 1165.2 ± 283.1 μg/ml, which was slightly reduced by 10 nm GNPs (888.4 ± 206.3 μg/ml) and 50 nm GNPs (799.1 ± 61.3 μg/ml ) on day 5 (Table 1); however this reduction was not statistically significant (ANVOA F=0.784, P=0.549). The A260/A280 ratio for the RNA extracted from liver samples ranged from 2.034 to 2.112, indicating sufficient purity of extracted RNA (Table 1). The total RNA yield from the control kidney was 143.6 ± 16.8 μg/ml, which was significantly reduced (ANOVA F=3.983, P=0.016) by 50 nm GNPs (58.2 ± 12.1 μg/ml) (Table 2). The A260/A280 ratio for the RNA extracted from kidney samples ranged from 2.078 to 2.122, indicating sufficient purity of extracted RNA from kidneys (Table 2). Figure 1 shows a comparative view of total RNA yield from liver and kidney samples.
| Group | RNA yield (µg/ml) | A280 | A260 | A260/A280 |
|---|---|---|---|---|
| Control | 1165.2 ± 283.1 | 14.26 ± 3.39 | 29.12 ± 7.07 | 2.034 ± 0.003 |
| GNP10, Day 1 | 1030.4 ± 110.1 | 12.21 ± 1.36 | 25.72 ± 2.75 | 2.112 ± 0.001 |
| GNP10, Day 5 | 888.4 ± 206.3 | 10.70 ± 2.49 | 22.16 ± 5.15 | 2.070 ± 0.003 |
| GNP50, Day 1 | 1153.1 ± 170.7 | 13.84 ± 2.12 | 28.82 ± 4.27 | 2.090 ± 0.001 |
| GNP50, Day 5 | 799.1 ± 61.3 | 9.53 ± 0.61 | 19.92 ± 1.53 | 2.084 ± 0.002 |
Table 1: Total RNA extraction from liver of rats treated with gold nanoparticles.
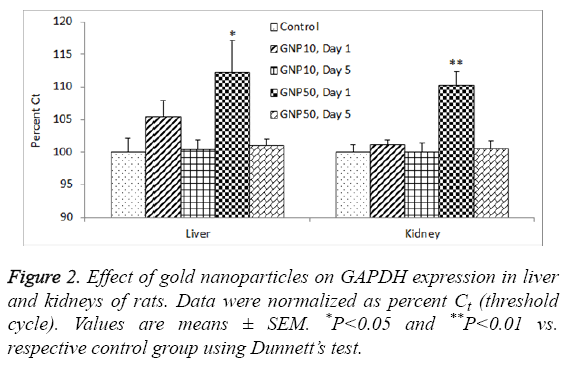
There was a significant reduction in GAPDH gene expression (high Ct value) in livers of rats treated with 50 nm GNPs, day 1 post-dosing (ANOVA F=3.799, P=0.02). The same group of rats also showed significant reduction in GAPDH expression in kidneys as well (ANOVA F=3.986, P=0.001) (Figure 2). In livers, the total RNA levels were not correlated with GAPDH expression (R=0.013, P=0.950) whereas in kidneys, there was a significant correlation between total RNA and GAPDH expression (R=-0.739, P=0.001).
Discussion
Our results showed several folds higher RNA yield from liver as compared to kidneys of rats (Figure 1). Liver is the major organ where most of the metabolic activities take place resulting in massive turnover of mRNA of a large number of genes. On the other hand, kidney is mainly an excretory organ with the involvement of comparatively less number of genes expressions. With the RNeasy procedure, all RNA molecules longer than 200 nucleotides are purified. The procedure provides enrichment for mRNA since most RNAs<200 nucleotides (such as 5.8S rRNA, 5S rRNA and tRNAs, which together comprise 15-20% of total RNA) are selectively excluded from the lysate. Poor RNA yield from the kidney tissues may partly be due to the fact that we collected kidney tissue from the cortex region. Previously we have shown that kidney medulla has more intense gene expression as compared to kidney cortex [26].
Degradation of mRNA is an important component of gene function that controls the steady-state concentration of functional transcript levels in the cell [27]. Although majority of transcripts are stable, specific half-life of each mRNA is precisely related to its physiological role [28-30]. For instance, most of the housekeeping genes have longer mRNA half-lives whereas proteins that are required for a limited time in the cell often have mRNAs with shorter half-lives [31]. Sharova et al. [32] successfully determined the rate of mRNA decay for 19977 non-redundant genes by microarray analysis of RNA samples obtained from mouse embryonic stem cells. Median estimated half-life was 7.1 h and only<100 genes, including Prdm1, Myc, Gadd45 g, Foxa2, Hes5 and Trib1, showed halflife less than 1 h. In general, mRNA species with short half-life were enriched among genes with regulatory functions (transcription factors), whereas mRNA species with long halflife were enriched among genes related to metabolism and structure (extracellular matrix, cytoskeleton) [32].
Administration of GNPs did not affect the yield of RNA from liver (Table 1) whereas the 50 nm GNPs significantly reduced the yield of RNA from kidneys, on day 1 but not after day 5 (Table 2). This could be due to an acute phase response on the expression of some specific genes. Previous studies have shown an acute phage induction of proinflammatory cytokines by GNPs in rat liver and kidneys [23]. Earlier we have shown that the proinflammatory response on day 1 was comparatively more severe with 50 nm GNPs than 10 nm GNPs [24]. Particle size plays a key role in immunoreactivity because of the differential deposition of complement proteins that is affected by the size of nanoparticles. The complement system plays an important role in uptake and clearance of nanoparticles as well as the modulation of immune response [33]. Intraperitoneal injection of 10 nm diameter GNPs significantly increased liver malondialdehyde without altering glutathione levels in rat liver, on 3 and 7 days post-dosing [34]. Nanogold flakes ameliorated alcohol-induced liver injury by maintaining the hepatic antioxidative status [35].
| Group | RNA yield (µg/ml) | A280 | A260 | A260/A280 |
|---|---|---|---|---|
| Control | 143.6 ± 16.8 | 1.693 ± 0.201 | 3.591 ± 0.423 | 2.122 ± 0.005 |
| GNP10, Day 1 | 161.8 ± 22.5 | 1.925 ± 0.264 | 4.048 ± 0.563 | 2.102 ± 0.003 |
| GNP10, Day 5 | 120.1 ± 23.6 | 1.423 ± 0.281 | 2.994 ± 0.591 | 2.106 ± 0.005 |
| GNP50, Day 1 | 58.2 ± 12.1* | 0.697 ± 0.141* | 1.449 ± 0.298* | 2.078 ± 0.018 |
| GNP50, Day 5 | 134.2 ± 21.6 | 1.594 ± 0.261 | 3.355 ± 0.542 | 2.106 ± 0.008 |
Table 2: Total RNA extraction from kidney of rats treated with gold nanoparticles.
The results of GAPDH expression in kidneys (Figure 2) correlated with the total RNA yield from the same organ (Table 2). However, such a correlation was not observed in liver of rats. This could be due to sub-acute liver injury induced by GNPs leading to vacuolated swelling of the cytoplasm of the hepatocytes [36]. Impact of altered organ weight on RNA yield from organs of GNP-treated rats may be ruled out as previous studies failed to notice any change in liver and kidney weights of animals treated with GNPs. In rats, 14 days of repeated oral administration of 5-15 nm diameter GNPs (325-1300 μg/kg) did not produce any significant change in organo-somatic index (weight of organ/body weight × 100) of brain, kidney, intestine, liver, lung, spleen and stomach [37]. In mice, intraperitoneal administration of GNPs (1100 μg/kg for 28 days) did not affect the weight of various organs including liver and kidneys [38].
We used RNAlater technology for RNA stabilization, which is an absolute prerequisite for reliable gene expression analysis. Immediate stabilization of RNA in biological samples is important because, after harvesting the samples, alterations in gene expression pattern occur due to RNA degradation as well as due to transcriptional induction. These alterations significantly affect the accuracy of results and must be avoided for reliable gene expression analysis. The RNAlater technology is based on RNA stabilization reagent, which ensures immediate preservation of the gene expression pattern in animal tissues enabling reliable gene expression analysis. After harvesting, the respective tissues are immediately submerged in appropriate volume of RNA stabilization reagent for rapid permeation into tissues causing in-situ stabilization and protection of cellular RNA. According to manufacturer’s guidelines, the reagent preserves RNA for up to 1 day at 37°C, 7 days at room temperature and 4 weeks at 2-8°C, allowing transportation, storage, and shipping of samples without ice or dry ice.
Conclusion
In conclusion, the total RNA yield was several folds higher from liver as compared to kidneys of rats. Treatment of rats with 10 nm and 50 nm GNPs did not affect the RNA yield from liver whereas the 50 nm GNPs significantly reduced the RNA yield from the kidneys of rats. The expression of housekeeping gene, GAPDH, was significantly reduced in livers and kidneys of rats treated with 50 nm GNPs, day 1 post-dosing. Further studies are warranted to investigate the impact of GNPs exposure on RNA yield from different organs of rats.
Acknowledgements
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding the Research Group No. RGP-066.
References
- Weir A, Westerhoff P, Fabricius L, Hristovski K, von Goetz N. Titanium dioxide nanoparticles in food and personal care products. Environ Sci Technol 2012; 46: 2242-2250.
- Eswar KA, Rouhi J, Husairi FS, Dalvand R, Alrokayan SA, Khan HA, Mahmood R, Abdullah S. Hydrothermal growth of flower-like ZnO nanostructures on porous silicon substrate. J Mol Struc 2014; 1074: 140-143.
- Kang SH, Nafiujjaman M, Nurunnabi M, Li L, Khan HA, Cho KJ, Huh KM, Lee Y. Hybrid photoactive nanomaterial composed of gold nanoparticles, pheophorbide-A and hyaluronic acid as a targeted bimodal phototherapy. Macromol Res 2015; 23: 474-484.
- Tapia-Hernandez JA, Torres-Chavez PI, Ramirez-Wong B, Rascon-Chu A, Plascencia-Jatomea M, Barreras-Urbina CG, Rangel-Vazquez NA, Rodriguez-Felix F. Micro- and nanoparticles by electrospray: advances and applications in foods. J Agric Food Chem 2015; 63: 4699-4707.
- Zehedina K, Nurunnabi M, Nafiujjaman M, Reeck G, Khan HA, Cho KJ, Lee Y. A hyaluronic acid nanogel for photo-chemo theranostic of lung cancer with simultaneous light-responsive controlled release of doxorubicin. Nanoscale 2015; 7: 10680-10689.
- Nurunnabi M, Parvez K, Nafiujjaman M, Revuri V, Khan HA, Feng X, Lee Y. Bioapplication of graphene oxide derivatives: drug/gene delivery, imaging, polymeric modification, toxicology, therapeutics and challenges. RSC Advances 2015; 5: 42141-42161.
- Nafiujjaman M, Nurunnabi M, Kang SH, Reeck G, Khan HA, Lee Y. Ternary graphene quantum dot-polydopamine-Mn3O4 nanoparticles for optical imaging guided photodynamic therapy and T1-weighted magnetic resonance imaging. J Mater Chem B 2015; 3: 5815-5823.
- Zhang X, Qian J, Pan B. Fabrication of novel magnetic nanoparticles of multifunctionality for water decontamination. Environ Sci Technol 2016; 50: 881-889.
- Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett 2012; 12: 5192-5195.
- Guo P. The emerging field of RNA nanotechnology. Nat Nanotechnol 2010; 5: 833-842.
- Khan HA, Shanker R. Toxicity of nanomaterials. Biomed Res Int 2015; 2015: 521014.
- Panchapakesan B, Book-Newell B, Sethu P, Rao M, Irudayaraj J. Gold nanoprobes for theranostics. Nanomedicine (Lond) 2011; 6: 1787-1811.
- Pissuwan D, Niidome T, Cortie MB. The forthcoming applications of gold nanoparticles in drug and gene delivery systems. J Control Release 2011; 149: 65-71.
- Shaat H, Mostafa A, Moustafa M, Gamal-Eldeen A, Emam A. Modified gold nanoparticles for intracellular delivery of anti-liver cancer siRNA. Int J Pharm 2016; 504: 125-133.
- Watanabe K, Ohtsuki T. Photocontrolled intracellular RNA delivery using nanoparticles or carrier-photosensitizer conjugates. Prog Mol Biol Transl Sci 2016; 139: 101-119.
- Huang W, Liu Z, Zhou G, Tian A, Sun N. Magnetic gold nanoparticle-mediated small interference RNA silencing Bag-1 gene for colon cancer therapy. Oncol Rep 2016; 35: 978-984.
- Jackson SR, Wong AC, Travis AR, Catrina IE, Bratu DP. Applications of hairpin DNA-functionalized gold nanoparticles for imaging mRNA in living cells. Methods Enzymol 2016; 572: 87-103.
- Yang WJ, Li XB, Li YY, Zhao LF, He WL. Quantification of microRNA by gold nanoparticle probes. Anal Biochem 2008; 376: 183-188.
- Grzincic EM, Yang JA, Drnevich J, Falagan-Lotsch P, Murphy CJ. Global transcriptomic analysis of model human cell lines exposed to surface-modified gold nanoparticles: the effect of surface chemistry. Nanoscale 2015; 7: 1349-1362.
- Lai TH, Shieh JM, Tsou CJ, Wu WB. Gold nanoparticles induce heme oxygenase-1 expression through Nrf2 activation and Bach1 export in human vascular endothelial cells. Int J Nanomedicine 2015; 10: 5925-5939.
- Sharma M, Salisbury RL, Maurer EI, Hussain SM, Sulentic CE. Gold nanoparticles induce transcriptional activity of NF- κB in a B-lymphocyte cell line. Nanoscale 2013; 5: 3747-3756.
- Doss CG, Debajyoti C, Debottam S. The impact of gold nanoparticles on hTERT gene expression leading to termination of malignant tumor. Gene 2012; 493: 140-141.
- Khan HA, Abdelhalim MA, Alhomida AS, Al Ayed MS. Effects of naked gold nanoparticles on proinflammatory cytokines mRNA expression in rat liver and kidney. Biomed Res Int 2013; 2013: 590730.
- Khan HA, Abdelhalim MA, Alhomida AS, Al Ayed MS. Transient increase in IL-1ß, IL-6 and TNF-a genes expression in liver of rats exposed to gold nanoparticles. Genet Mol Res 2013; 12: 5851-5857.
- Khan HA, Ibrahim KE, Khan A, Alrokayan SH, Alhomida AS, Lee YK. Comparative evaluation of immunohistochemistry and real-time PCR for measuring proinflammatory cytokines gene expression in livers of rats treated with gold nanoparticles. Exp Toxicol Pathol 2016; 68: 381-390.
- Khan HA, Ibrahim KE, Khan A, Alrokayan SH, Alhomida A. Immunostaining of proinflammatory cytokines in renal cortex and medulla of rats exposed to gold nanoparticles. Histol Histopathol 2016: 11825.
- Raghavan A, Bohjanen PR. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief Funct Genomic Proteomic 2004; 3: 112-124.
- Yang E, van Nimwegen E, Zavolan M. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 2003; 13: 1863-1872.
- Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 2004; 15: 303-313.
- Newbury SF. Control of mRNA stability in eukaryotes. Biochem Soc Trans 2006; 34: 30-34.
- Hollams EM, Giles KM, Thomson AM, Leedman PJ. MRNA stability and the control of gene expression: implications for human disease. Neurochem Res 2002; 27: 957-980.
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MSH. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res 2009; 16: 45-58.
- Pondman KM, Pednekar L, Paudyal B, Tsolaki AG, Kouser L, Khan HA, Shamji MH, Haken BT, Stenbeck G, Sim RB, Kishore U. Innate immune humoral factors, C1q and factor H, with differential pattern recognition properties, alter macrophage response to carbon nanotubes. Nanomedicine 2015; 11: 2109-2118.
- Khan HA, Abdelhalim MA, Al-Ayed MS, Alhomida AS. Effect of gold nanoparticles on glutathione and malondialdehyde levels in liver, lung and heart of rats. Saudi J Biol Sci 2012; 19: 461-464.
- Chen YL, Peng HC, Tan SW, Tsai CY, Huang YH. Amelioration of ethanol-induced liver injury in rats by nanogold flakes. Alcohol 2013; 47: 467-472.
- Abdelhalim MA, Abdelmottaleb Moussa SA. The gold nanoparticle size and exposure duration effect on the liver and kidney function of rats: In vivo. Saudi J Biol Sci 2013; 20: 177-181.
- Jo MR, Bae SH, Go MR, Kim HJ, Hwang YG, Choi SJ. Toxicity and biokinetics of colloidal gold nanoparticles. Nanomaterials 2015; 5:835-850.
- Zhang XD, Wu HY, Wu D, Wang YY, Chang JH. Toxicologic effects of gold nanoparticles in vivo by different administration routes. Int J Nanomedicine 2010; 5: 771-781.