Research Article - Journal of RNA and Genomics (2016) Volume 12, Issue 1
The inhibition of Cg2076, the GHITM homologue in neurons of Drosophila Melanogaster can be rescued by Buffy
P Githure M’Angale and Brian E Staveley*
1Department of Biology, Memorial University of Newfoundland, 232 Elizabeth Avenue, St. John’s, Newfoundland and Labrador, Canada
Received Date: 01 July 2016; Revised Date: 01 August 2016; Accepted Date: 28 September 2016; Published Date: 03 October 2016
Copyright: © First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Abstract
Growth hormone-inducible transmembrane protein (GHITM) is an inner mitochondrial membrane protein that contains the Bax inhibitor-1 motif and is implicated in the regulation of mitochondrial morphology and especially cristae structure. The downregulation of GHITM results in fragmented mitochondria and the release of cytochrome c, while its upregulation delays the release of cytochrome c. We inhibited CG2076 the Drosophila GHITM homologue in the neurons using RNA interference and analysed the phenotypic consequences of this mitochondrial protein. The directed expression of GHITM-RNAi in neurons under the control of the Dopa decarboxylase (Ddc) transgene results in shortened lifespan and impaired climbing ability. The co-expression of Buffy, the only anti-apoptotic B cell lymphoma 2 (Bcl-2) protein in Drosophila, along with GHITM-RNAi results in suppression of the shortened lifespan and premature age-dependent loss in climbing ability. The inhibition of GHITM in the Drosophila eye results in decreased ommatidia number and elevated disruption of the ommatidial array, phenotypes that are rescued upon overexpression of Buffy. The inhibition of the mitochondrial located GHITM in the Ddc-Gal4-expressing neurons of Drosophila results in shortened lifespan and loss in climbing ability, phenotypes that are manifest of degeneration and death of dopaminergic neurons, and are improved upon overexpression of the pro-survival Buffy.
Keywords
CG2076, growth hormone-inducible transmembrane protein, Buffy, neurons, drosophila, transmembrane bax inhibitor-1 motif
Introduction
The transmembrane Bax inhibitor-1 motif containing (TMBIM) family consists of several antiapoptotic members that are evolutionarily conserved, being found in viruses, bacteria, protozoans, plants and animals (Hu et al, 2009; Rojas-Rivera and Hetz, 2015). This group of proteins is so diversely conserved that they are present in organisms where the Bcl- 2 family of proteins have not yet been identified. Generally, 6 members or orthologues can be present in an organism, that include TMBIM1/RECS1, TMBIM2/LFG, TMBIM3/ GRINA, TMBIM4/GAAP, TMBIM5/GHITM and TMBIM6/ BI-1 (Rojas-Rivera and Hetz, 2015). The different members are localised to different cellular organelles, with TMBIM1/ RECS1 predominantly found in the endosomal/lysosomal membranes; TMBIM2/LFG at the plasma and intracellular membranes of the Golgi and the endoplasmic reticulum (ER); TMBIM3/GRINA is primarily located at the ER and Golgi compartments; TMBIM4/GAAP to the Golgi apparatus and the ER; TMBIM5/GHITM to the mitochondrial inner membrane; and TMBIM6/BI-1 at the ER (Hu et al, 2009; Reimers et al, 2008; Lisak et al, 2015; Rojas-Rivera and Hetz, 2015). Growth hormone-inducible transmembrane protein (GHITM)/TMBIM5 also referred to as Mitochondrial morphology and cristae 1 (MICS1) is a mitochondrial inner membrane protein that is involved in mitochondria morphology and specifically the cristae and is implicated in the release of cytochrome c from the mitochondria (Oka et al, 2008). It was named GHITM as it was found dysregulated in expression analysis of inter-capsular brown adipose tissue of mice that were expressing a growth hormone antagonist (Li et al, 2001). This protein consists of seven transmembrane domains with a presequence as shown by the presence of a cleavage site at the amino (N)-terminal and is ubiquitously expressed in mammals (Yoshida et al, 2006; Reimers et al, 2007). GHITM/MICS1 regulates cell death by the regulation of mitochondria morphology, since the knock down of this gene results in mitochondrial fragmentation and cristae disorganization followed by the release of mitochondrial proapoptotic proteins that include the apoptogenic cytochrome c (Oka et al, 2008). Overexpression of GHITM/MICS1 is able to directly block the release of cytochrome c from the inner mitochondria membrane independent of Bax-induced permeabilization though it does not block apoptotic death. Its maintenance of mitochondrial morphology therefore, is distinct from its role in the apoptotic process.
The Drosophila melanogaster homologue is predicted to be CG2076 and CG1287 (Rojas-Rivera and Hetz, 2015), the two putative genes have 56% and 53% protein sequence identity to the human GHITM as determined by BLAST; CG2076 is more closely related to the human homologue. Bioinformatic studies establish that the two genes are very closely related, 67% identity and 82% similarity in their protein sequences. CG2076 has two annotated transcripts on FlyBase but only one is unique, while CG1287 has only one annotated transcript (Attrill et al, 2016). Drosophila has been used as a model organism to study the phenotypic consequences of differential gene expression and to model human diseases with very promising results (Staveley, 2014). DA neurons are sensitive to subtle differences in gene products and degenerate in an age-dependent manner which can be quantified by scoring climbing ability of the affected flies. We investigated the outcome of the inhibition of CG2076, the Drosophila melanogaster homologue of GHITM in the Ddc- GAL4-expressing neurons, and further determined whether the Bcl-2 proteins, known to be the ?guardians? of the mitochondria can rescue the CG2076/GHITM-induced phenotypes by the overexpression of the sole pro-survival Bcl-2 homologue in Drosophila, Buffy.
Material And Methods
Bioinformatic analysis
The protein sequences for Drosophila melanogaster: NP_610824.1, Homo sapiens: NP_036438.2, Xenopus tropicalis: NP_001072357.1, and Mus musculus: NP_082500.2 were sourced from National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/protein/). The functional domains were identified using the NCBI Conserved Domain Database (CDD) (Marchler-Bauer et al, 2015) (http://www. ncbi.nlm.nih.gov/cdd) and the Eukaryotic Linear Motif (Dinkel et al, 2016) (http://elm.eu.org/) which focuses on annotation and detection of eukaryotic linear motifs (ELMs), also known as short linear motifs (SLiMs). A Clustal Omega multiple sequence alignment (http://www.ebi.ac.uk/Tools/msa/clustalo/) (Goujon et al, 2010, Sievers et al, 2011) was used to show conservation of the Bax inhibitor-1 domain. Transmembrane domains were confirmed using TMpred (Artimo et al, 2012), a program based on statistical analysis of TMbase that identifies membrane-spanning regions (http://www.ch.embnet.org/ software/TMPRED_form.html). Further analysis of protein domains was performed with Phyre2 (Kelley et al, 2015), a web portal for protein modelling, prediction and analysis (http:// www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The sub-cellular and mitochondrial targeting signal was identified using TargetP (Emanuelsson et al, 2000) (http://www.cbs.dtu. dk/services/TargetP/) and MultiLoc2 (Blum et al, 2009) (https:// abi.inf.uni-tuebingen.de/Services/MultiLoc2).
Drosophila media and stocks
Stocks and crosses were maintained on standard cornmeal/ molasses/yeast/agar media treated with propionic acid and methylparaben. Aliquots of media were poured into plastic vials, allowed to solidify, and refrigerated at between 4ºC and 6ºC. Stocks were raised at room temperature while crosses and experiments for analysis of ageing and climbing ability were carried out at 25ºC while those for the eye analysis were performed at 29ºC. The CG2076 stock, w1118; P{GD3308}v5537 hereby referred to as UAS-GHITM-RNAi was obtained from the Vienna Drosophila Resource Center. UAS-Buffy (Quinn et al, 2003) was kindly provided by Dr. L. Quinn of University of Melbourne and Ddc-Gal4 flies (Li et al, 2000) by Dr. J. Hirsch of University of Virginia. GMR-Gal4 (Freeman, 1996) and UAS-lacZ flies were obtained from the Bloomington Drosophila Stock Center.
Drosophila derivative lines
The UAS-Buffy/CyO; Ddc-Gal4 and UAS-Buffy/CyO; GMRGal4 complex lines were used to overexpress Buffy in neurons and the developing eye and were produced employing standard homologous recombination and marker selection methods as previously described (M?Angale and Staveley, 2016a, M?Angale and Staveley, 2016c). Gel electrophoresis was used to confirm recombination events via the presence of a PCR product.
Ageing assay
Flies were aged using a standard protocol as previously described (Todd and Staveley, 2012; M?Angale and Staveley, 2016). Briefly, more than two hundred flies were aged per genotype and scored every two days for presence of deceased adults (Staveley et al, 1990). Longevity data was analysed using GraphPad Prism version 5.04 and survival curves were compared using the Log-rank (Mantel-Cox) test. Significance was determined at 95%, at a P-value less than or equal to 0.05 with Bonferroni correction.
Climbing assay
The climbing assay was performed as previously described (Todd and Staveley, 2004). Climbing indices were computed and then analysed using GraphPad Prism version 5.04. The 5-climbing index is a model generated for graded climbing analysis using non-linear regression and 5 is the highest level the flies can climb. Confidence intervals were compared at 95% at a P-value of 0.05.
Scanning electron microscopy of the drosophila eye
Crosses for the analysis of the Drosophila eye were made of each genotype at 29o C and a batch of male flies collected and assessed using a standard protocol previously described (M?Angale and Staveley, 2016). 10 different scanning electron micrographs of each genotype were analysed using the National Institutes of Health (NIH) ImageJ software (Schneider et al, 2012) and biometric analysis performed using GraphPad Prism version 5.04. The area of disruption of the ommatidial array was determined as detailed previously (M?Angale and Staveley, 2012). Statistical comparisons were evaluated using unpaired student T-tests. P-values less than 0.05 were considered significant.
Results
Human GHITM/MICS1 is closely related to Drosophila CG2076
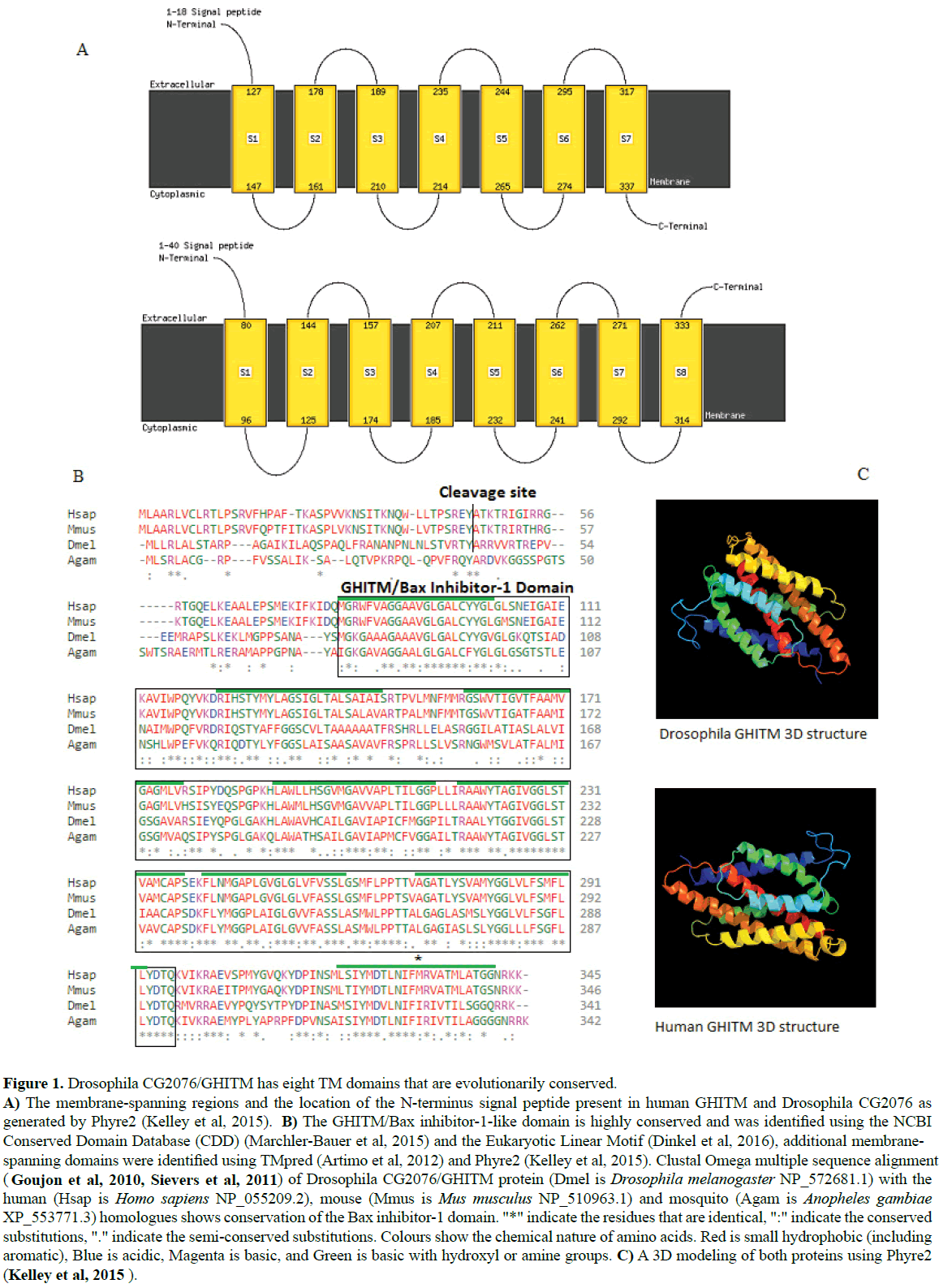
Human GHITM/MICS1 is closely related to Drosophila CG2076 The Drosophila CG2076, the human GHITM homologue contains the Growth hormone-inducible transmembrane hormone domain that is closely related to the Bax inhibitor-1- like superfamily as determined by NCBI Conserved Domain Database Search (CDD) (Marchler-Bauer et al, 2015). CG2076 is composed of 341 amino acids and shows 56% identity and 73% similarity to the 345 amino acids human GHITM (Figure 1). The Drosophila homologue has eight TM domains (Figure 1A) and the human transcript has seven TM domains as determined by the Eukaryotic linear motif (ELM) resource search (Dinkel et al, 2016) and Phyre2 (Kelley et al, 2015). A prediction of membrane-spanning regions using TMpred (Artimo et al, 2012), indicates that all the aligned sequences possesses eight TM domains, the first seven fall within the GHITM/Bax inhibitor- 1-like domain and the eighth membrane-spanning region falls outside the protein family domain. A similar search using Phyre2 (Kelley et al, 2015), gave similar results for CG2076 but returned seven TM domains for the human version. A multiple sequence alignment of protein sequences using Clustal Omega (Goujon et al, 2010; Sievers et al, 2011) shows high conservation of the Bax inhibitor-1-like domain (Figure 1B). CG2076/GHITM is localised to the mitochondria and has a mitochondrial targeting peptide with a presequence cleavage site at amino acid 43 as predicted using TargetP (Emanuelsson et al, 2000) and MultiLoc (Blum et al, 2009). A 3D modelling of both proteins using Phyre2 (Kelley et al, 2015) is shown (Figure 1C).
Figure 1:Drosophila CG2076/GHITM has eight TM domains that are evolutionarily conserved. A) The membrane-spanning regions and the location of the N-terminus signal peptide present in human GHITM and Drosophila CG2076 as generated by Phyre2 (Kelley et al, 2015). B) The GHITM/Bax inhibitor-1-like domain is highly conserved and was identified using the NCBI Conserved Domain Database (CDD) (Marchler-Bauer et al, 2015) and the Eukaryotic Linear Motif (Dinkel et al, 2016), additional membranespanning domains were identified using TMpred (Artimo et al, 2012) and Phyre2 (Kelley et al, 2015). Clustal Omega multiple sequence alignment ( Goujon et al, 2010, Sievers et al, 2011) of Drosophila CG2076/GHITM protein (Dmel is Drosophila melanogaster NP_572681.1) with the human (Hsap is Homo sapiens NP_055209.2), mouse (Mmus is Mus musculus NP_510963.1) and mosquito (Agam is Anopheles gambiae XP_553771.3) homologues shows conservation of the Bax inhibitor-1 domain. "*" indicate the residues that are identical, ":" indicate the conserved substitutions, "." indicate the semi-conserved substitutions. Colours show the chemical nature of amino acids. Red is small hydrophobic (including aromatic), Blue is acidic, Magenta is basic, and Green is basic with hydroxyl or amine groups. C) A 3D modeling of both proteins using Phyre2 (Kelley et al, 2015 ).
Inhibition of CG2076/GHITM in the Ddc-GAL4-expressing neurons shortens lifespan and impairs climbing ability
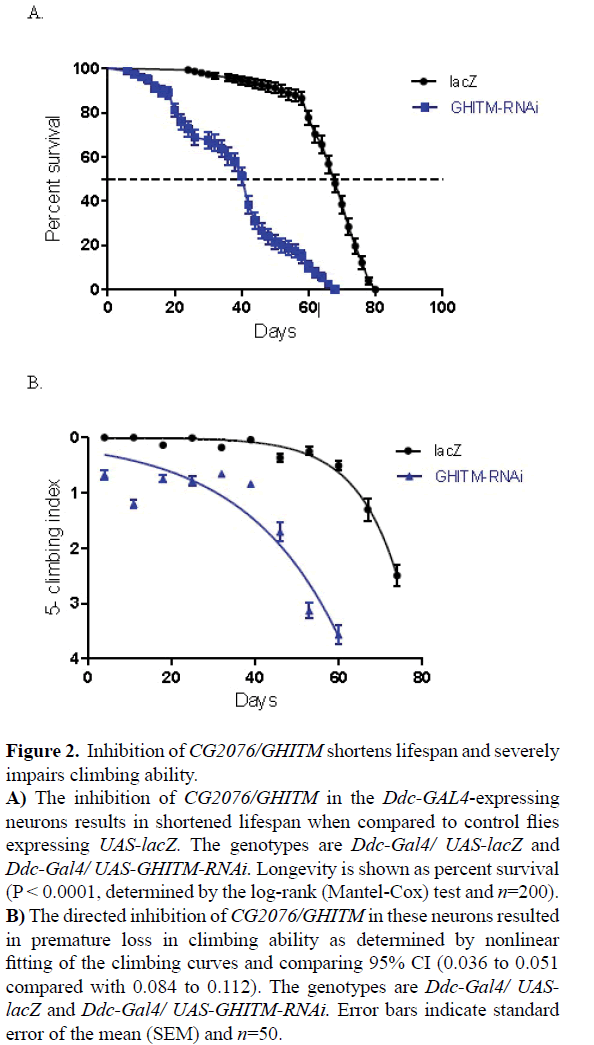
The suppression of CG2076/GHITM in the DA neurons results in severely shortened lifespan and highly impaired climbing ability. The median survival of GHITM-RNAi flies was 42 days compared to 68 days for the controls that express the benign lacZ as determined by Log-rank (Mantel-Cox) test (Figure 2A). The directed inhibition of CG2076/GHITM in the Ddc-GAL4- expressing neurons produces flies with significantly impaired climbing ability as determined by a nonlinear fit of the climbing curves (Figure 2B). The comparison of the confidence intervals (CI) at 95% indicate a significant difference between the GHITM-RNAi flies with 0.036 to 0.051 compared with 0.084 to 0.112 for the controls. These results suggest that CG2076/ GHITM is required for the normal function of these neurons in Drosophila.
Figure 2: Inhibition of CG2076/GHITM shortens lifespan and severely impairs climbing ability.
A) The inhibition of CG2076/GHITM in the Ddc-GAL4-expressing neurons results in shortened lifespan when compared to control flies expressing UAS-lacZ. The genotypes are Ddc-Gal4/ UAS-lacZ and Ddc-Gal4/ UAS-GHITM-RNAi. Longevity is shown as percent survival (P < 0.0001, determined by the log-rank (Mantel-Cox) test and n=200).
B) The directed inhibition of CG2076/GHITM in these neurons resulted in premature loss in climbing ability as determined by nonlinear fitting of the climbing curves and comparing 95% CI (0.036 to 0.051 compared with 0.084 to 0.112). The genotypes are Ddc-Gal4/ UASlacZ and Ddc-Gal4/ UAS-GHITM-RNAi. Error bars indicate standard error of the mean (SEM) and n=50.
Buffy suppresses the loss of CG2076/GHITM-induced phenotypes
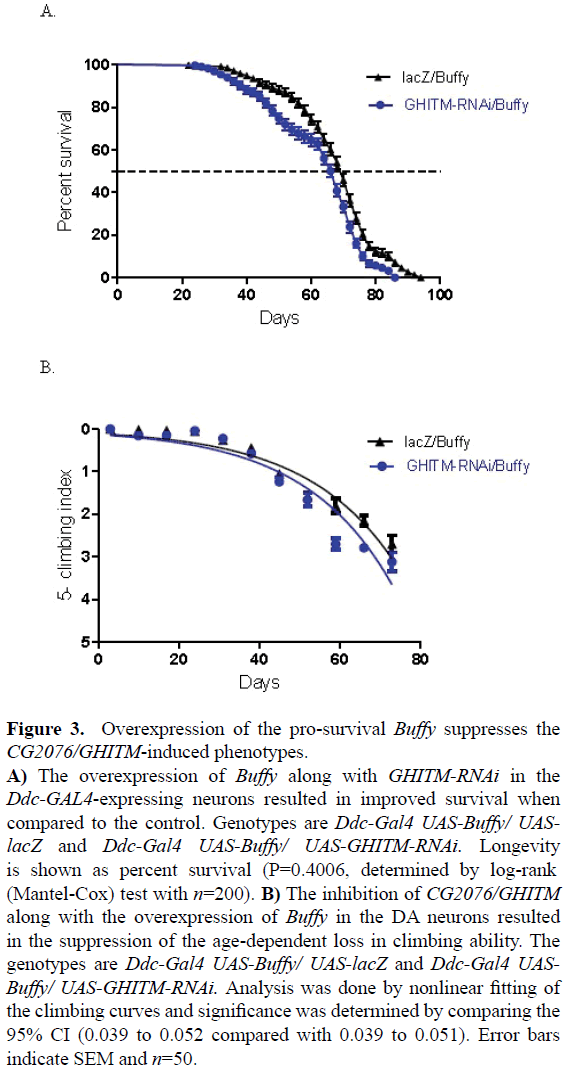
The overexpression of the pro-survival Bcl-2 homologue Buffy along with the suppression of CG2076/GHITM in the Ddc-GAL4-expressing neurons results in a significant increase in lifespan and improved climbing ability. The coexpression of Buffy with GHITM-RNAi results in increased median survival of 68 days when compared to Buffy control flies with a median survival of 72 days as determined by Logrank test (Figure 3A). The climbing ability of the GHITMRNAi flies was improved as determined by comparison of the climbing curves at 95% CI with 0.039 to 0.052 compared with 0.039 to 0.051 which was not significant (Figure 3B). These results suggest a pro-survival role for Buffy; it increases the general ?healthspan? of GHITM-RNAi flies as it improves survival and locomotor function when CG2076/ GHITM is inhibited in these neurons.
Figure 3: Overexpression of the pro-survival Buffy suppresses the CG2076/GHITM-induced phenotypes.
A) The overexpression of Buffy along with GHITM-RNAi in the Ddc-GAL4-expressing neurons resulted in improved survival when compared to the control. Genotypes are Ddc-Gal4 UAS-Buffy/ UASlacZ and Ddc-Gal4 UAS-Buffy/ UAS-GHITM-RNAi. Longevity is shown as percent survival (P=0.4006, determined by log-rank (Mantel-Cox) test with n=200).
B) The inhibition of CG2076/GHITM along with the overexpression of Buffy in the DA neurons resulted in the suppression of the age-dependent loss in climbing ability. The genotypes are Ddc-Gal4 UAS-Buffy/ UAS-lacZ and Ddc-Gal4 UASBuffy/ UAS-GHITM-RNAi. Analysis was done by nonlinear fitting of the climbing curves and significance was determined by comparing the 95% CI (0.039 to 0.052 compared with 0.039 to 0.051). Error bars indicate SEM and n=50.
Inhibition of CG2076/GHITM in the eye decreases ommatidia number and increases disruption, phenotypes that are rescued upon Buffy overexpression
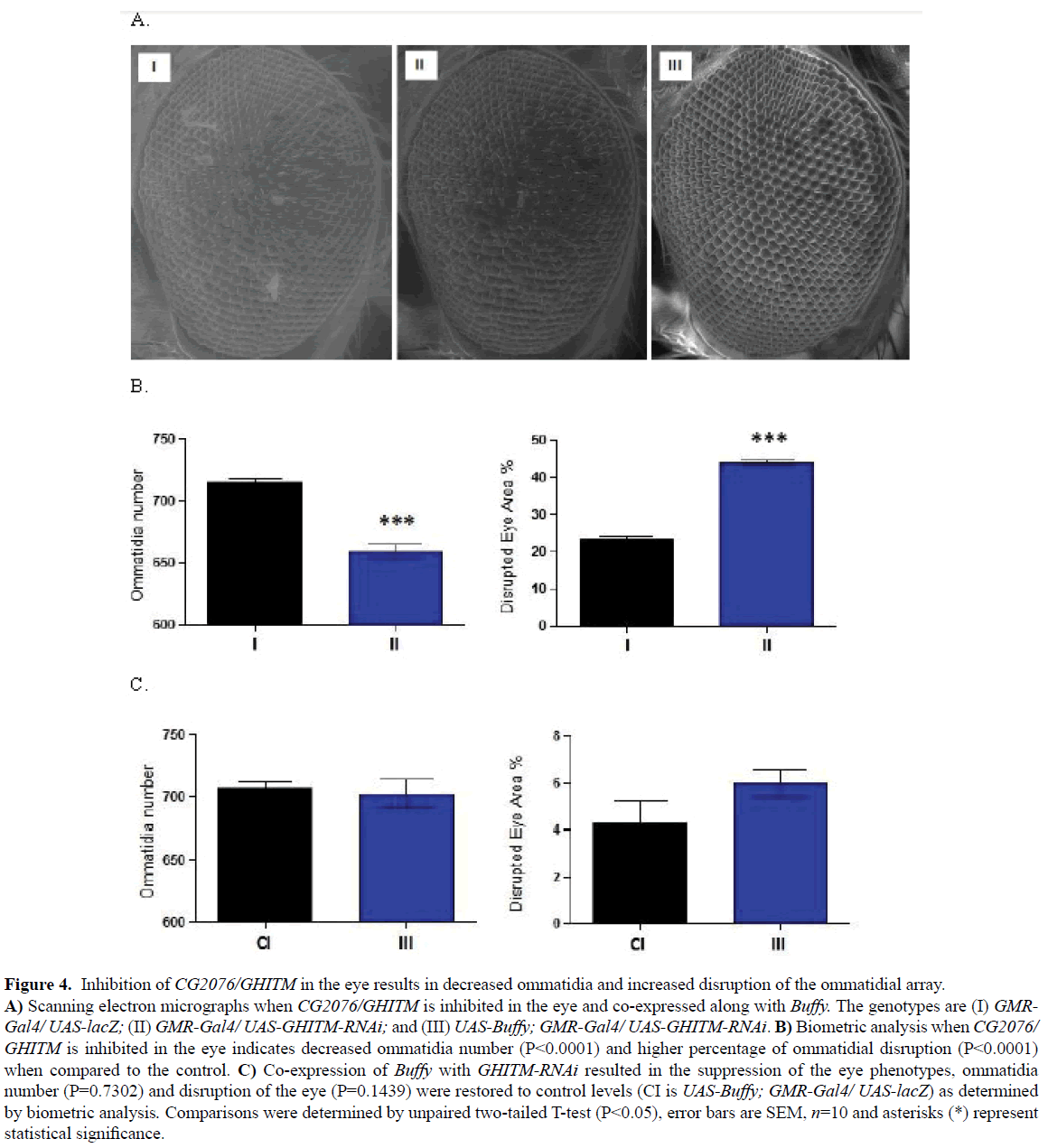
The inhibition of CG2076/GHITM in the eye under the direction of the GMR-Gal4 transgene decreases ommatidia number and results in significant disruption of the ommatidial array (Figures 4A, II and 4B) as determined by an unpaired T-test p< 0.0001. The overexpression of Buffy along with the inhibition of CG2076/ GHITM restored the number of ommatidia and the percentage disruption to control levels as determined by an unpaired T-test, p>0.50 (Figures 4A, III and 4C). Taken together, these results suggest that CG2076/GHITM may play a developmental role in the Drosophila eye and that Buffy suppresses the developmental eye defects that result from its inhibition.
Figure 4: Inhibition of CG2076/GHITM in the eye results in decreased ommatidia and increased disruption of the ommatidial array. A) Scanning electron micrographs when CG2076/GHITM is inhibited in the eye and co-expressed along with Buffy. The genotypes are (I) GMRGal4/ UAS-lacZ; (II) GMR-Gal4/ UAS-GHITM-RNAi; and (III) UAS-Buffy; GMR-Gal4/ UAS-GHITM-RNAi. B) Biometric analysis when CG2076/ GHITM is inhibited in the eye indicates decreased ommatidia number (P< 0.0001) and higher percentage of ommatidial disruption (P< 0.0001) when compared to the control. C) Co-expression of Buffy with GHITM-RNAi resulted in the suppression of the eye phenotypes, ommatidia number (P=0.7302) and disruption of the eye (P=0.1439) were restored to control levels (CI is UAS-Buffy; GMR-Gal4/ UAS-lacZ) as determined by biometric analysis. Comparisons were determined by unpaired two-tailed T-test (P< 0.05), error bars are SEM, n=10 and asterisks (*) represent statistical significance.
Discussion
The precise function of the BI-1 consensus motif (UPF0005) is not known, but it encodes six to seven transmembrane spanning domains that are highly conserved in many species and signifies an important biological function (Reimers et al, 2007). Bioinformatic analysis of protein sequences from previous work (Rojas-Rivera and Hetz, 2015) and our own study showed CG2076 and CG1287 to be the strongest candidates for Drosophila GHITM; CG2076 appears to be the closest with a sequence identity of 56% and 73% similarity, though this does not exempt CG1287, our main consideration was the high degree of similarity to human GHITM as determined by BLAST. Therefore, we propose that CG2076 is the Drosophila homologue of GHITM/TMBIM5.
The inhibition of CG2076/GHITM using RNA interference under the direction of the Ddc-Gal4 transgene in the dopaminergic neurons of Drosophila resulted in decreased median survival and severely impaired climbing ability. The general wellbeing of these flies was highly compromised as demonstrated by the shortened lifespan and premature retardation in climbing ability. GHITM is a mitochondria inner membrane protein that possesses an N-terminal presequence that is important for its expression (Yoshida et al, 2006; Reimers et al, 2007). The presence of a mitochondria targeting sequence firmly localizes it to the mitochondria, while the loss of GHITM function induces cell death (Oka et al, 2008). This cell death has been attributed to the fragmentation of the mitochondria and the subsequent release of the apoptogenic factor cytochrome c. In addition, GHITM was reported to be physically associated with cytochrome c. Dopaminergic neurons are sensitive to energy requirements and mitochondrial dysfunction is the main culprit that leads to their degeneration and death (Ryan et al, 2015). The integrity of the mitochondria is vital to the survival of these important motor neurons and any disruption in their function is implicated in disease including Parkinson disease (Rugarli and Langer, 2012; Franco-Iborra et al, 2015; Ryan et al, 2015). The results obtained suggest a strong role for CG2076/ GHITM in neuroprotection in Drosophila since the loss of function in the Ddc-Gal4-expressing neurons results in shortened lifespan and impaired climbing ability. The observed GHITM-induced cell death is possibly through mitochondrial dysfunction.
The pro-survival Bcl-2 family of proteins are known to protect the mitochondria from breach by the pro-apoptotic members and releasing a variety of apoptogenic molecules that include cytochrome c (Siddiqui et al, 2015). The sole pro-survival Bcl-2 homologue in Drosophila is Buffy (Quinn et al, 2003), and the overexpression of Buffy along with the inhibition of CG2076/GHITM resulted in the suppression of the CG2076/ GHITM-induced phenotypes. The overexpression of GHITM partially blocks the release of cytochrome c during apoptosis (Oka et al, 2008), while its downregulation induced a failure to maintain normal mitochondrial network and disorganization of the cristae. We have previously shown that Buffy rescues Ddc-GAL4-expressing neurons when co-expressed with the neurotoxic a-synuclein (M?Angale and Staveley, 2016), or the pro-apoptotic Debcl (M?Angale and Staveley, 2016a), or the Parkinson disease related High temperature requirement A2 (HtrA2) (M?Angale and Staveley, 2016b). This Buffy protection may be induced by pro-survival pathways though it is possible that Buffy regulates mitochondrial cell death as the phenotypes that result from the inhibition of CG2076/GHITM are rescued by Buffy. This in addition highlights the protective role of CG2076/ GHITM in the neurons as its phenotypes can be rescued by the overexpression of the pro-survival Buffy.
The inhibition of CG2076/GHITM in the Drosophila eye under the direction of the GMR-Gal4 transgene results in a depressed number of ommatidia, this was mostly due to the fusion of the ommatidia and the extensive ommatidial disarray. The inhibition of CG2076/GHITM in the Drosophila eye seems to exacerbate the Gal4-induced apoptosis that manifests as roughened eye phenotype (Kramer and Staveley, 2003). The overexpression of Buffy along with the inhibition of CG2076/GHITM results in the suppression of the Gal4 and the Gal4 in addition to the GHITM-RNAi phenotypes, with the number of ommatidia and the degree of roughened eye restored to control levels. Buffy seems to ameliorate this phenotype possibly via a general action on survival signals through the mitochondria or through a concerted function to rescue GHITM-induced apoptosis at the mitochondria.
Conclusion
CG2076 appears to be the GHITM/TMBIM5 homologue in Drosophila based on sequence homology, the presence of a mitochondria targeting signal, and a 43 amino acids presequence, features that it strongly shares with the human GHITM transcript. The inhibition of CG2076/GHITM in the Ddc-GAL4-expressing neurons of Drosophila results in a severely shortened lifespan and an age-dependent loss in climbing ability, phenotypes that are strongly associated with the degeneration and loss of DA neurons, and may as well point to a novel model of Parkinson disease. The overexpression of the pro-cell survival Buffy along with the inhibition of CG2076/ GHITM results in the rescue of the observed phenotypes
Acknowledgements
PGM was partially funded by Department of Biology Teaching Assistantships and a School of Graduate Studies Fellowship from Memorial University of Newfoundland. BES was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant. The funding bodies were not involved in the design of the study and in the collection, analysis, and interpretation of the data and writing the manuscript.
Competing Interests
The authors declare that there are no competing interests.
List of Abbrevations
Ddc: dopa decarboxylase GHITM: growth hormone-inducible transmembrane protein GMR: glass multiple reporter RNAi: ribonucleic acid interference SEM: standard error of the mean TMBIM: transmembrane Bax inhibitor 1 motif
References
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, et al. 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597-W603.
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, et al. 2016. FlyBase: establishing a gene group resource for drosophila melanogaster. Nucleic Acids Res. 44, D786-D792.
- Blum T, Briesemeister S and Kohlbacher O. 2009. MultiLoc2: integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 10, 274.
- Dinkel H, Van Roey K, Michael S, Kumar M, Uyar B, et al. 2016. ELM 2016-data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 44, D294-D300.
- Emanuelsson O, Nielsen H, Brunak S and von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. JMol Biol. 300, 1005-1016.
- Franco-Iborra S, Vila M and Perier C. 2015. The parkinson disease mitochondrial hypothesis: where are we at? Neuroscientist. 22, 266-277.
- Freeman M. 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 87, 651-660.
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J and Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL?EBI. Nucleic Acids Res. 38, W695-W699.
- Hu L, Smith TF and Goldberger G. 2009. LFG: a candidate apoptosis regulatory gene family. Apoptosis. 14, 1255-1265.
- Kelley LA, Mezulis S, Yates CM, Wass MN and Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols. 10, 845-858.
- Kramer JM and Staveley BE. 2003. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Gen Mol Res. 2, 43-47.
- Li H, Chaney S, Roberts IJ, Forte M and Hirsh J. 2000. Ectopic G-protein expression in dopamine and serotonin neurons blocks cocaine sensitization in Drosophila melanogaster. Curr Biol. 10, 211-214.
- Li Y, Kelder B and Kopchick JJ. 2001. Identification, isolation, and cloning of growth hormone (GH)-inducible interscapular brown adipose complementary deoxyribonucleic acid from GH antagonist mice. Endocrinology. 142, 2937-2945.
- Lisak DA, Schacht T, Enders V, Habicht J, Kiviluoto S, et al. 2015. The transmembrane Bax inhibitor motif (TMBIM) containing protein family: Tissue expression, intracellular localization and effects on the ER CA(2)(+)-filling state. Biochim Biophys Acta. 1853, 2104-2114.
- M'Angale PG and Staveley BE. 2012. Effects of a-synuclein expression in the developing Drosophila eye. Dros Info Serv. 95, 85-89.
- M'Angale PG and Staveley BE. 2016. The Bcl-2 homologue Buffy rescues alpha-synuclein-induced Parkinson disease-like phenotypes in Drosophila. BMC Neurosci. 17, 24.
- M?Angale PG and Staveley BE. 2016a. Bcl-2 homologue Debcl enhances a-synuclein-induced phenotypes in Drosophila PeerJ. 4, e2461.
- M?Angale PG and Staveley BE. 2016b. The HtrA2 Drosophila model of Parkinson Disease is suppressed by the pro-survival Bcl-2 Buffy. Genome. In Press.
- M?Angale PG and Staveley BE. 2016c. Inhibition of Atg6 and Pi3K59F autophagy genes in neurons decreases lifespan and locomotor ability in Drosophila melanogaster. Gen Mol Res. In Press.
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222-D226.
- Oka T, Sayano T, Tamai S, Yokota S, Kato H, Fujii G and Mihara K. 2008. Identification of a novel protein MICS1 that is involved in maintenance of mitochondrial morphology and apoptotic release of cytochrome c. Mol Biol Cell. 19, 2597-2608.
- Quinn L, Coombe M, Mills K and Daish T. 2003. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 22, 3568-3579.
- Reimers K, Choi CY, Bucan V and Vogt PM. 2007. The growth-hormone inducible transmembrane protein (GHITM) belongs to the Bax inhibitory protein-like family. Int J Biol Sci. 3, 471-476.
- Reimers K, Choi CY, Bucan V and Vogt PM. 2008. The Bax Inhibitor-1 (BI-1) family in apoptosis and tumorigenesis. Curr Mol Med. 8, 148-156.
- Rojas-Rivera D and Hetz C. 2015. TMBIM protein family: ancestral regulators of cell death. Oncogene. 34, 269-280.
- Rugarli EI and Langer T. 2012. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 31, 1336-1349.
- Ryan BJ, Hoek S, Fon EA and Wade-Martins R. 2015. Mitochondrial dysfunction and mitophagy in Parkinson's: from familial to sporadic disease. Trends Biochem Sci. 40, 200-210.
- Schneider CA, Rasband WS and Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9, 671-675.
- Siddiqui WA, Ahad A and Ahsan H. 2015. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch Toxicol. 89, 289-317.
- 30">Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7, 539.
- Staveley BE. 2014. Drosophila models of parkinson disease. In MS LeDoux ed. Movement Disorders: Genetics and Models. Elsevier Science. 345-354.
- Staveley BE, Phillips JP and Hilliker AJ. 1990. Phenotypic consequences of copper-zinc superoxide dismutase overexpression in Drosophila melanogaster. Genome. 33, 867-872.
- Todd AM and Staveley BE. 2004. Novel assay and analysis for measuring climbing ability in Drosophila. Dros Info Serv. 87, 101-107.
- Todd AM and Staveley BE. 2012. Expression of Pink1 with alpha-synuclein in the dopaminergic neurons of Drosophila leads to increases in both lifespan and healthspan. Genet Mol Res. 11, 1497-1502.
- Yoshida T, Nagata S and Kataoka H. 2006. GHITM is an ortholog of the Bombyx mori prothoracic gland-derived receptor (Pgdr) that is ubiquitously expressed in mammalian cells and requires an N-terminal signal sequence for expression. Biochem Biophys Res Commun. 341, 13-18.