Research Article - Biomedical Research (2018) Volume 29, Issue 12
Sub-acute exposure effect of selected polycyclic aromatic hydrocarbons on protein levels of epigenetic modifiers in non-cancerous hepatic model
Sitlali del Rosario Olguín-Reyes, Sandra Luz Hernández-Ojeda, Tzipe Govezensky, Rafael Camacho- Carranza, Jesús Javier Espinosa-Aguirre*Department of Genomic Medicine and Environmental Toxicology, Institute of Biomedical Research, National Autonomous University of Mexico, University City, Coyoacán, México
- *Corresponding Author:
- Jesús Javier Espinosa-Aguirre
Department of Genomic Medicine and Environmental Toxicology
Institute of Biomedical Research
National Autonomous University of Mexico
University City, Coyoacán, México
Accepted date: May 24, 2018
DOI: 10.4066/biomedicalresearch.29-18-342
Visit for more related articles at Biomedical ResearchAbstract
It has been proposed that in the first stage of carcinogenesis, the progenitor cell may have epigenetics changes before acquisition of mutations, driving the cell to acquire malignant characteristics. Some polycyclic aromatic hydrocarbons (PAH) are carcinogenic compounds, like benzofluorene, benzo (a) pyrene, 3-methylcholanthrene, and 7, 12-dimethylbenzanthracene, which can also produce such mutations, therefore, that epigenetic changes may be favoured in the presence of these molecules. Here, normal rat liver epithelial cell cultures were exposed in a sub-acute way to these selected PAH in order to evaluate changes in the level of proteins involved in the modification of epigenetic changes, such as epigenetic modifiers proteins. In spite that these compounds are structurally related, have similar metabolism pathways and are ligands of same nuclear receptor, they exhibit a different pre-mutational insult to DNA. In consequence, the results show that at least BaP, BF and DMBA modified with a different pattern the protein levels of epigenetic modifiers proteins.
Keywords
Polycyclic aromatic hydrocarbons, Global DNA methylation, DNA methyltransferases, Histone deacetylases, SIRT1, 5-aza-2-deoxycytidine.
Introduction
Polycyclic Aromatic Hydrocarbons (PAH) are anthropogenic compounds found in several sources, such as cigarette smoke, coal tar, charcoal cooked food, to mention a few [1]. They display a wide range of modes of action for toxicity, such as interaction and activation of nuclear receptors, causing repression or activation of gene transcription. Despite that, they are best known as mutagenic and carcinogenic compounds, because their metabolites produce covalent bonds with DNA, which can become in fixed mutations through cell divisions [2]. An example of such covalent bonds between DNA and PAH is given by population living in industrialized areas which present elevated PAH-DNA adduct levels in their blood leukocytes [3-5].
Previously, it has been proposed that cumulative mutations will eventually arise to the initiating mutation in the progenitor cell, allowing to carcinogenesis, but even when the prototypical PAH mutagen benzo (a) pyrene (BaP) has 53.1 × 10-8 relative adduct levels in normal human fibroblast cell culture exposed to 10 μM for 72 h, it achieves as much as 0.92% rate of mutation [6], while in its epithelial-mesenchymal transition model it takes 1 μM BaP 24 weeks to present cell transformation [7]. Thus, although DNA damage occurs fast, loss of cell identity takes months of exposure to the carcinogenic compound.
Additionally, some studies proved that epigenetic changes are acquired by non-mutated normal tissues under chemical exposure from which tumor arises, such an example is provided by mutations in p53, which is a marker for lung carcinogenesis [8]. Establishment of mutations on p53 is a late event in lung carcinogenesis, while DNA methylation changes are observed at early stages of this process [9]. It is proposed that epigenetic changes in a progenitor cell precede the initiating mutation [10,11]. Thus, efforts must be done to determine changes in epigenetic processes, as well as changes in the modifiers of those processes, at early time exposure to mutagenic agents in order to determine how they are improving the establishment of progenitor cell.
Epigenetic processes modulate gene expression along life time, they are inherited, involved in the development of several diseases, and can be modulated by environment [12]. Among the proteins that govern epigenetic mechanisms the main groups are DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) [13]. The former group is involved in DNA methylation, which is a mode of gene regulation characterized by methylation at C-5 position of cytosine in CpG dinucleotides, being a marker for either recruitment or prevention of transcriptional factors as well as modifiers of chromatin for gene repression. The latter are involved in histone deacetylation reaction, which play a critical role in formation of a euchromatin state, therefore facilitating gene expression, additionally, aside from histone proteins, they also catalyze deacetylation reactions at other kind of proteins [14].
DNMTs and HDACs are designated as epigenetic modifiers by the tumor progenitor theory. This theory suggests that the structure of chromatin suffers changes at very early stages in the cancer process and involves epigenetic modulators, modifiers and mediators, rather than simply mutations as an initiation mechanism for carcinogenesis. Epigenetic modifiers are those proteins involved directly in alteration of DNA methylation, chromatin remodeling or structure of chromatin, such as DNMTs and HDACs. These epigenetic modifiers lay their actions over epigenetic mediators, which are genes or proteins that can drive tumors or its progenitor cells towards a neoplastic state, like the insulin like growth factor 2 or WNT signaling members [10,13].
Epigenetic modifiers and mediators can be modulated by processes like cellular signaling and metabolic pathways, which are classified as epigenetic modulators, examples are p53 and RAS signaling. In addition, cellular status, like inflammation, oxidant stress or injury by environmental chemicals, biological and physical agents, can affect the action of epigenetic modifiers and modulators [10,13]. Thus, the present study aimed to profile protein level changes of selected epigenetic modifiers after exposure to four selected PAH compounds.
To address our aim, we selected four well known PAH carcinogens that have been studied thoroughly for several years, BaP, 3-methylcholanthrene (3MC), dimethylbenz (a) anthracene (DMBA), and benzofluorene (BF). These compounds are classical substrates and inducers of cytochrome P450 enzymes (CYP), particularly the isoforms CYP1B1 and CYP1A1 [15], which in turn generate highly reactive metabolites, which react with proteins and DNA, leading to mutagenesis and carcinogenesis [16]. In addition to these effects, they are estrogenic compounds, modulators of cell cycle, proliferation and even modulators of angiogenesis [17-20].
Some studies conducted in cancerous cell cultures showed that BaP can modify DNA methylation and protein levels of DNMTs. Although different mechanisms of action were suggested, DNA hypomethylation was a constant result among those studies, while the effect of BaP over DNMTs protein level was not the same among those studies [21,22]. Actually, it is proposed that cancer cells exhibited global DNA hypomethylation at early mutagen/carcinogen exposure [23], thus, experimental studies must be performed in non-cancerous cells in order to determine early epigenetic changes by carcinogenic compounds in cells with an epigenome nearly unaltered. Then, to reach our experimental purposes, we chose the normal rat liver cell line C9 (C9 cells) because it is metabolically active and have similar epigenetic status to hepatocytes primary cultures [24]. There are very few reports addressing the PAH effects on DNMTs, HDACs, or any other epigenetic modifier protein in hepatic cells. Indirect studies have been performed to make a relationship between PAH exposure and DNA methylation changes in specific genes such as c-Myc proto-oncogene and RRSSF1A tumor suppressor gene, which are involved in human hepatocellular carcinoma (HCC) establishment. In this cellular model, c-Myc is overexpressed according to its DNA hypomethylation, while RRSSF1A mRNA and protein are inversely related to its DNA hypermethylation, and those conditions are related with the increase of PAH-adducts [25-27]. In another hand, a study conducted in murine hepatoma cancer cells exposed to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) showed that this compound induces cyp1a1 promoter DNA hypomethylation, consequently TCDD increases gene expression of this cytochrome 3 to 5 times in subsequent dioxin exposures. Surprisingly, the DNA demethylation of the cyp1a1 promoter by TCDD require d the recruitment of the epigenetic modifiers proteins Tet2 and Tet3, which are enzymes involved in active DNA demethylation achieved through the conversion of 5- methyl group of CpG into hydroxymethyl moiety that is recognized by thymidine DNA glycosylase, base excision repair, and then replaced with an unaltered cytosine [28]. These studies support the hypothesis that PAHs are able to change epigenetic modifiers proteins conferring a cell environment suitable for the establishment of neoplastic cell.
As was reported for BaP in cancerous cell models, 3MC was also able to alter DNMT1 protein level and DNA methylation, but in a multistage model for lung carcinogenesis [29,30]. Thus, we selected the DNMT protein family to be profiled after C9 cell culture exposure to BaP, BF, 3MC and DMBA. We also determined if the protein levels of these epigenetic modifiers can be translated into alteration of global DNA methylation, which was determined via detection of the immunofluorescence mark for 5-methylcytosine.
The other main group of enzymes belonging to the epigenetic modifiers group are HDACs. They have been implicated in the DNA damage response, homologous recombination, and chromatin integrity [31]. Increased activity of these enzymes may allow loss of acetylation, particularly loss of global monoacetylation at histone 4, which is common in human tumor cells [23]. Furthermore, in vitro and in vivo studies reported changes in HDAC1 and HADC2 protein levels after exposure to cigarette smoke and 3MC [32,33]; also, even when the studies were not related to PAH exposure, hepatic models have been studied to find the role that HDACs are playing in liver cancer process. From those studies, HDAC1, 2, 3, 5, 6 were found to be involved in anti-apoptotic and proliferative processes in liver cancer cell lines [34-41]. Therefore, we selected some HDAC proteins to be profiled after C9 cell exposure to the four selected PAH mutagens.
Materials and Methods
Reagents and antibodies
Trypsin, dimethyl sulfoxide (DMSO), BaP, 3MC, DMBA, BF, 5-aza-2-deoxycytidine (5AzadC), was obtained from Sigma Aldrich Co. (St. Louis, MO, USA). Alexa Fluor 488® secondary antibody was acquired from Invitrogen-Life Technologies (San Francisco, CA, USA). Antibodies against CYP1B1, DNMT1, DNMT3a, HDAC1, HDAC2, HDAC3 and HDAC4 were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). Antibody against DNMT3b and SIRT1 was obtained from Abcam (San Francisco, CA, USA). Antibody against GAPDH was obtained from GeneTex (Irvin, CA, USA). HRP-conjugated antibody against rabbit was acquired from Millipore (Billerica, MA, USA). Dulbecco's modified Eagle media (DMEM) and antibiotic-anti-mycotic were obtained from Gibco-Life Technologies (San Francisco, CA, USA). Rat liver epithelial cell line Clone 9, CRL-1439, was supplied by ATCC (Manassas, VA, USA). Fetal bovine serum (FBS) was purchased from ByProductos (Guadalajara, Jalisco, Mexico).
Cell culture
We selected the normal rat liver epithelial cell line Clone 9 (C9 cells) as a representative cell line of normal rat hepatocytes cells. C9 cells were grown in DMEM supplemented with 10% FBS and 0.1% antibiotic-antimycotic at 37°C and 5% CO2. 5 × 10-5 C9 cells were plated at dilution 1:10 in 100 mm petri dishes, media was renewed every 3 d, 100% confluence cells were harvested with 0.25% trypsin-1mM EDTA. All experiments were performed after five passages of the C9 cells. Experimental treatments were performed by plating 5 × 105 C9 cells on 100-mm petri dishes at 1:10 dilution with daily media changes. The PAH compounds were dissolved in 0.1% v/v DMSO and final concentrations of 10 M were added to media culture after 48 h of C9 cells plating and continued for 48 h with media renewal each 24 h. 0.1% DMSO was used as vehicle control. C9 cells achieved ~100% confluence at the end of treatments.
To have a positive control for DNA hypomethylation and to determine that changes in DNMTs and DNA methylation were related, we included an experiment where C9 cells were exposed to the DNMTs inhibitor 5-aza-2-deoxycytidine [42]. 0.5 μM of this compound was added into media after 24 h of C9 cells plating and continued for 72 h, alone or in combination with BaP for last 48 h. At the end of the exposure time, C9 cells were harvested and stored at -80°C for future analysis or processed at the moment for protein and immunofluorescence assays. The results represent the average of 3 to 6 independent assays.
Western blot
25 μg of total C9 cell lysate proteins were resolved using a 7.5% polyacrylamide gel under denatured conditions. Resolved proteins were transferred to a nitrocellulose membrane at 250 mA for 3 h at 4°C. Membranes were blocked with 3% m/v BSA-TBS for 2 h at 4°C, exposed to primary antibodies at 1:500 dilution in 0.1% m/v BSA-TBS overnight at 4°C, and secondary antibodies at 1:10000 dilution in 0.1% m/v BSATBS at room temperature for 1 h [43]. To detect the protein of interest, membranes were exposed to AmershamTM ECL Prime Western Blotting Detection Reagent. Digital images were obtained with a Kodak Gel Logic 200 Imaging System and analyzed with the NIH ImageJ software program [44].
5mC immunofluorescence
Immunofluorescence (IF) assays were performed as reported by Liu et al. [45]. Briefly, C9 cells were fixed for 5 min with 4% paraformaldehyde, permeabilized with 0.5% Triton-PBS, and blocked with 15% foetal rabbit serum. Later, C9 cells were exposed to mouse antibodies against 5mC overnight at 4°C.
After this, C9 cells were exposed to rabbit Alexa Fluor® 488 anti-mouse antibodies at room temperature for 2 h, and mounted on glass slides with DAPI-VectaShield (Vector Laboratories, Burlingame, CA, USA). Fluorescence was detected using a fluorescence microscope Olympus BX51-WI Microscope and analyzed with NIH ImageJ software program [44].
Statistical analysis
Data obtained from image analysis of immunoblots for each treatment was compared against vehicle control using one-way ANOVA and Dunnett’s multiple comparisons as post hoc test.
Data obtained from immunofluorescence assays were analyzed by comparing each treatment against each other by one-way ANOVA and Tukey’s multiple comparisons as post hoc test. Data reported in figures are the mean ± standard error. Statistical significance was established to be P<0.05. Analyses were performed with SAS 9.0.
Results
To evaluate the epigenetic profile of the PAH compounds in an in vitro non-cancerous model we chose the normal liver epithelial cell line C9. In this model, we determined that the PAH compounds selected were not cytotoxic at concentrations below 50 μM.
Based on this and on similar studies where concentrations of 2.5 to 40 μM of these PAH were used [21,22], we decided to expose C9 cell culture to 10 μM each PAH compound for 48 h.
To assess the protein profile level of DNMT proteins after exposure to BF, BaP, 3MC and DMBA, we determined protein levels of DNMT1, DNMT3a and DNMT3b in total cell protein.
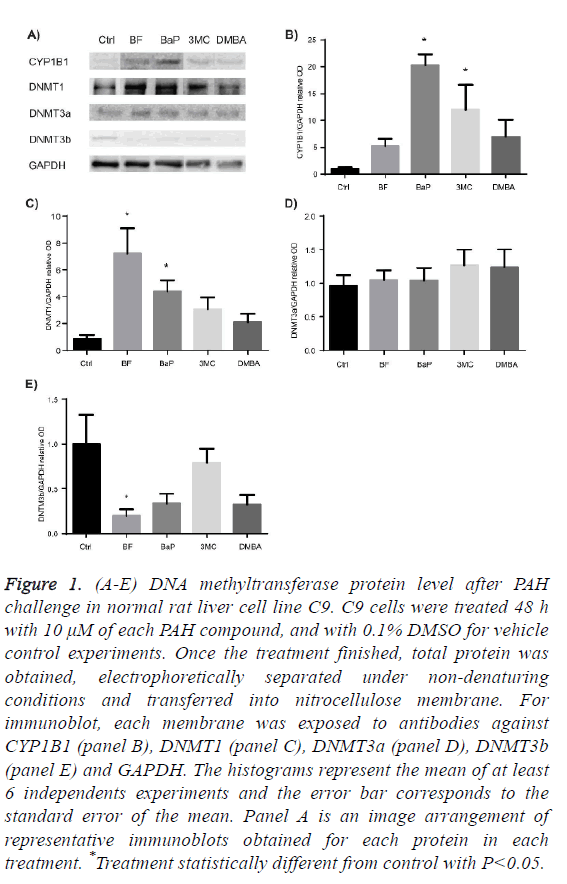
Figure 1A-1C shows that only BF and BaP significantly increased the protein level of DNMT1 enzyme but at different extent.
Figure 1D shows that the protein level of DNMT3a in the presence of each PAH did not exhibit statistical difference from control, while the protein level of DNTM3b was significantly diminished after exposure to BF, as Figure 1E shows.
Figure 1: (A-E) DNA methyltransferase protein level after PAH challenge in normal rat liver cell line C9. C9 cells were treated 48 h with 10 μM of each PAH compound, and with 0.1% DMSO for vehicle control experiments. Once the treatment finished, total protein was obtained, electrophoretically separated under non-denaturing conditions and transferred into nitrocellulose membrane. For immunoblot, each membrane was exposed to antibodies against CYP1B1 (panel B), DNMT1 (panel C), DNMT3a (panel D), DNMT3b (panel E) and GAPDH. The histograms represent the mean of at least 6 independents experiments and the error bar corresponds to the standard error of the mean. Panel A is an image arrangement of representative immunoblots obtained for each protein in each treatment. *Treatment statistically different from control with P<0.05.
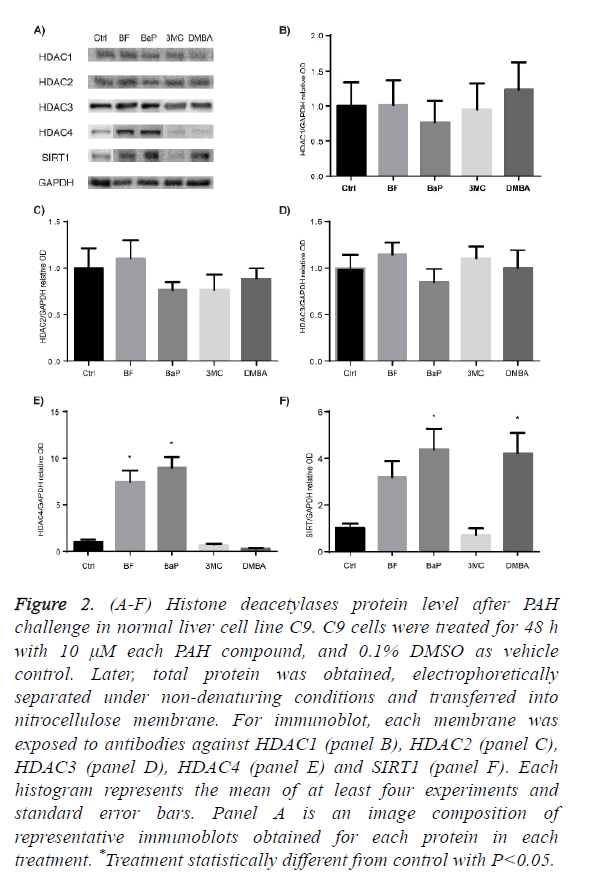
HDAC proteins family are divided into 4 classes, thus, we selected HDAC1, HDAC2 and HDAC3 from class I HDACs, HDAC4 from class II, and SIRT1 from class III, to determine their protein level profile after C9 cell culture exposure to BaP, BF, DMBA, and 3MC.
Figures 2A to 2D show that none of the evaluated PAH affected the protein levels of HDAC1, HDAC2 and HDAC3, whereas Figure 2E shows that HDAC4 was significantly increased after exposure to BF and BaP. Figure 2F shows that SIRT1 was significantly increased by BaP and DMBA, and BF increased it, although without statistical difference from control.
Figure 2: (A-F) Histone deacetylases protein level after PAH challenge in normal liver cell line C9. C9 cells were treated for 48 h with 10 μM each PAH compound, and 0.1% DMSO as vehicle control. Later, total protein was obtained, electrophoretically separated under non-denaturing conditions and transferred into nitrocellulose membrane. For immunoblot, each membrane was exposed to antibodies against HDAC1 (panel B), HDAC2 (panel C), HDAC3 (panel D), HDAC4 (panel E) and SIRT1 (panel F). Each histogram represents the mean of at least four experiments and standard error bars. Panel A is an image composition of representative immunoblots obtained for each protein in each treatment. *Treatment statistically different from control with P<0.05.
Summarizing, among the DNMTs, DNMT1 was modulated with significance by BF and BaP, and DNMT3b was diminished by BF. Among HDACs tested, HDAC 4 and SIRT1 were modulated; HDAC4 was increased by BF and BaP; and SIRT1 was increased by the BaP and DMBA.
Under the conditions of this study, results suggested that BF and BaP have major effects on the modulation of epigenetic modifiers being DNMT1, HDAC4 and SIRT1 up-regulated.
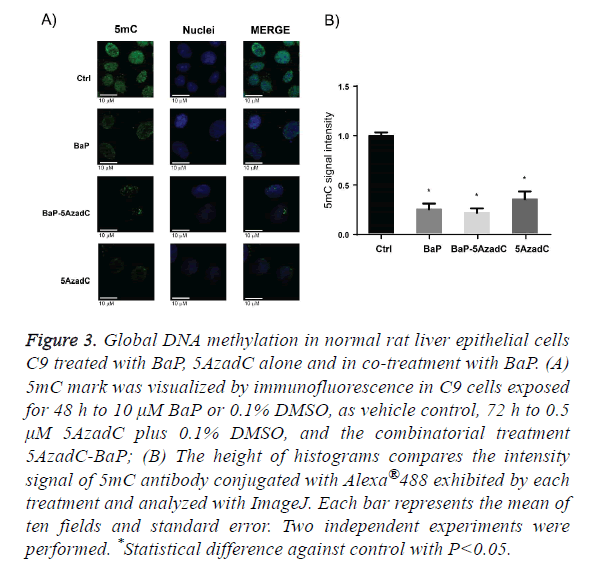
BaP as the prototypical compound of the PAH chemical group, it is the most studied and well know PAH compound. Consequently, we decided to evaluate global DNA methylation in C9 cells after exposure to BaP to explore whether the changes on DNMT1 protein levels by this PAH are translating in changes on DNA methylation. To address this aim we employed same scheme of treatment of BaP as in the profiling of epigenetic modifiers protein level, and signal intensity for 5mC antibody conjugated with Alexa 488 was evaluated. Figure 3A shows that the C9 cell culture exposure to BaP displayed a statistically global DNA hypomethylation state and this change in DNA methylation, as Figure 3B shows. The DNA hypomethylation resulting from exposure of C9 cells to BaP does not agree with DNMTs protein levels obtained after PAH exposure. Thus, to obtain more insight of these data, we evaluated the global DNA methylation after 72 h C9 cell culture exposure to 5AzadC, a DNMTs inhibitor. Likewise, we wondered if DNMTs inhibition by 5AzadC could be affected by the increased protein levels of DNMT1 due to BaP exposure, being reflected on DNA methylation. Then, we pretreated C9 cell culture 24 h with 5AzadC and this exposure was continued for 48 h by 5AzadC in co-treatment with BaP. From this experimental design, we obtained that exposure to 5AzadC achieved low levels of global DNA methylation and they were not statistically different from that obtained after BaP treatment; the same was obtained with C9 cell culture exposure to 5AzadC-BaP combination.
Figure 3: Global DNA methylation in normal rat liver epithelial cells C9 treated with BaP, 5AzadC alone and in co-treatment with BaP. (A) 5mC mark was visualized by immunofluorescence in C9 cells exposed for 48 h to 10 μM BaP or 0.1% DMSO, as vehicle control, 72 h to 0.5 μM 5AzadC plus 0.1% DMSO, and the combinatorial treatment 5AzadC-BaP; (B) The height of histograms compares the intensity signal of 5mC antibody conjugated with Alexa®488 exhibited by each treatment and analyzed with ImageJ. Each bar represents the mean of ten fields and standard error. Two independent experiments were performed. *Statistical difference against control with P<0.05.
Discussion
There is much knowledge about mutagenic properties of PAHs and now it is known that mutation is a late event in the process called initiation of carcinogenesis [8]. It has been proposed that cancer arises along three steps: epigenetic disruption of progenitor cells, initiating mutation, and genetic and epigenetic plasticity [13]. Therefore, carcinogenic compounds, such as PAHs, may have a role in the epigenetic disruption of progenitor cells before the establishment of key mutations. It is of our interest to know whether selected PAH compounds may exhibit epigenetic disruption properties at early time after their exposure. The tumor progenitor theory also proposes the involvement of epigenetic modifiers, modulators and mediators as players in the process of carcinogenesis. Among the main epigenetic modifiers are two families of proteins, DNMTs and HDACs [13].
In eukaryotes, DNMT family includes three proteins: the de novo DNMT3a and DNMT3b, which are involved in the methylation of un-methylated DNA; and the de novo and maintenance DNMT1 that is involved in DNA methylation of hemi-methylated DNA, thus perpetuates DNA methylation marks after each cell cycle [46].
There is evidence that DNMTs protein levels are frequently increased in human tumors [47-50], and also it was observed a progressive increase of protein levels of DNMT1 and DNMT3a along lung carcinogenesis induction by 3MC/ diethynitrosamine [29], as well as progressive loss of global DNA methylation [30]. Therefore, we explore whether four selected PAH can alter the protein levels of DNMTs in a non-cancerous model. Our results show that PAH modulate DNMTs protein levels in a differential extent: they increased the protein levels of DNMT1, but only BF and BaP achieved statistical significance. The increase of DNMT1 protein levels after BF and BaP treatment obtained in this study could be a response to DNA damage due to the accumulation of cells in phase S of cell cycle in order to repair the damage in DNA produced [51].
Another explanation for the increase of DNMT1 in C9 cell culture after exposure to BaP and BF is given by the results from a study conducted in HeLa cell culture exposed to BaP. In this study was found that this compound promotes the destruction of the N-terminal portion of DNMT1 which prevents it to go to proteasome degradation [9,52]. This lead us to think that the increase of DNMT1 protein levels by BF and BaP also could be related to the accumulation of this protein due to the prevention of its degradation, but this matter remains to be resolved through more studies in non-cancerous cells as well as in initiation stage of carcinogenesis.
As we mentioned before, loss of DNA methylation was also reported as an early event in the carcinogenesis multistage model, therefore, we determined the global DNA methylation status of the normal rat liver hepatocyte C9 cell culture after exposure to BaP. The results show that the exposure to this compound leads to the loss of global DNA methylation, which agrees with studies in cancer progression models where similar observations were made [29,30,53-56]. The fact that BaP exhibits a global DNA hypomethylation effect in noncancerous cells strongly supports the idea that epigenetic disruption would happen at initial stages of carcinogenesis development.
Additionally, global DNA hypomethylation exhibited by BaP could be a result from DNA damage response because it is known that poly (ADP-ribose) (PAR) moiety is increased during this event and this moiety is also an inhibitor of DNMT1 catalytic activity either as a free molecule or by binding to poly (ADP-ribose) polymerase 1 (PARP1) [57]. Some studies conducted in 16HBEC cells showed that global DNA hypomethylation induced by BaP is related to increased levels of PAR [22,58]. Also, as we mentioned before, loss of DNA methylation at local regions in specific genes, such as oncogenic c-Myc gene or xenobiotic metabolizing cyp1a1 gene, which DNA hypo-methylated status is related with exposure to BaP and, at least for the last gene could be the way BaP is driving the loss of methyl moieties on DNA, is through passive DNA demethylation conducted by Tet2 and Tet3 epigenetic modifiers [28]. Then, even when other mechanisms allow the increase of DNMT1 protein levels, this suggests that DNA hypomethylation by PAHs would be a general mechanism for this kind of compounds which is displayed at early time exposure and that is maintained along carcinogenesis progression.
Since PAH treatments result in the increasing of DNMT1 protein levels as well as global DNA hypomethylation, we decided to explore how global DNA methylation behaves in C9 cells after a challenge with an inhibitor of DNMTs and a PAH compound at the same time. 5-aza-2-deoxycytidine (5AzadC) was chosen as DNMTs inhibitor [42,59] Its effect alone was tested and, as expected, it significantly diminished the levels of global DNA methylation. In addition, even when our results showed that BaP increases the protein level of DNMT1, under our experimental conditions, this compound did not change the level of DNA methylation achieved by 5AzadC, suggesting that this PAH also promotes the loss of DNMT1 activity or or acts through different mechanisms besides global loss of DNA methylation, probably through a passive DNA de-methylation [28].
The other important group of epigenetic modifiers proteins is the HDAC family, which is comprised of 4 groups of enzymes: group 1) HDAC 1, 2, 3 and 8; group 2) HDAC 4-7, 9 and 10; group 3) Sirtuins (SIRT) 1-7, classified into 4 groups; and group 4) HDAC 11 [60,61]. They are grouped based on their catalytic mechanism similarities; groups 1, 2 and 4 are zincdependent amidohydrolases, while group 3 is a NAD+- dependent deacetylase activity.
Cancer cells exhibit loss of acetylation of histone residues [23,41,61,62], which in turn suggests that histone deacetylases must be over-expressed in these kind of cells. In addition, several cancer types exhibits over-expression of HDACs, such is the case of prostate, gastric, colon, liver cancers [41,61] which over-express HDAC1 [63], HDAC2 [64-66] and HDAC3 [67]. Over-expression of HDACs protein levels will result in increased cell proliferation, loss of differentiation and evasion of apoptosis [61,62], features that improve an initiated cell to become a tumor cell. Then, if since the beginning of the exposure to a PAH mutagen it is modifying the HDAC proteins that affect several cellular pathways, such as DNA damage repair, then it can affect the cell environment for acquisition of the aforementioned features.
Class I, II and III HDACs have been implicated in the DNA damage response, homologous recombination and chromatin integrity [31], thus we wondered whether these proteins are affected after a sub-acute challenge with BF, BaP, 3MC and DMBA, which are promoters of DNA repair due to their covalently binding to DNA. We evaluated the protein levels of HDAC1, HDAC2 and HDAC3 after PAH sub-acute exposure, but none of these proteins were significantly altered, even when previous studies have reported the protein degradation of HDAC1, HDAC2 and HDAC3 after 4 h of exposure to cigarette smoke in alveolar cells [32], and in mouse cerebral vascular endothelial cells 3MC was also reported as an agent that causes up-regulation of HDAC1 [33]. Studies on this regard must be done considering a temporal effect of at least BaP and 3MC, which have been reported as inducers of changes in protein levels of class I HDACs.
From class II HDACs, we evaluated possible changes of HDAC4. This protein has emerged as key protein for the maintaining of the G2 cell cycle checkpoint [68]. HDAC4 shuttles from the cytoplasm to the nucleus following DNA damage [69]. In the present study, this protein was significantly increased by BF and BaP. It was suggested that its persistence is linked to an unsuccessful DNA repair [68], and also its overexpression has been reported in many cancer types [41,61,70].
Finally, SIRT1 protein levels were increased after sub-acute exposure to BaP and DMBA. This is an intriguing result because this protein has been reported to be both up and downregulated in cancer cells and, due to its over-expression in some tumors, it is called an oncogenic protein, but for its absence or diminished levels in cancer cells it is also called tumor suppressor [41,61,71]. The disruption of SIRT1 leads to increased apoptosis after DNA damage, although, disruption of its deacetylation capacity leads to a highly sensitized cells to stress damage response [72,73]. The increase of SIRT1 protein level after exposure to BF, BaP and DMBA may be a normal response after a proper DNA repair where SIRT1 increases in order to stabilize p53 to an inactive state [74]. However, all chronic stimuli like PAH exposure will finally lie in disruption of the protein, thus, it may lead SIRT1 permanently increased.
Conclusion
The obtained results showed that BF, BaP, 3MC and DMBA affected differentially the protein levels of the epigenetic modifiers DNMTs and HDACs. Furthermore, exposure to BaP leads C9 cells to go to a global DNA hypomethylation state, even when DNMT1 levels were increased. Our results show that at least BF and BaP are environmental agents that affect epigenetic modifiers at early time exposure in non-cancerous cells. Deep and thorough research must be made to decipher how early changes are orchestrated for these well-known carcinogens, and how these changes on DNMT1, HDAC4 and SIRT1 may improve the establishment of mutations in the cell tumor progenitor.
Since human hepatic c-Myc and RASSF1A and mouse hepatic cyp1a1 are sensitive to DNA methylation, it would be interesting to search if the mechanism involved in their DNA methylation status is somehow related with the changes achieved for DNMT1 after C9 cells exposure to BaP.
Also, it would be interesting to determine whether the changes observed on DNTM1, HDAC4 and SIRT1 protein levels are involved with proliferation, anti-apoptotic or loss of differentiation of epithelial cells derived from liver, and if they are prompting the acquisition of mutations that accelerate getting neoplastic features.
Finally, since lung cancer is the most common cancer related with PAHs exposure, it will be so interesting to determine if the results obtained in this hepatic model also will be obtained in normal lung epithelial cells.
Acknowledgment
The authors are grateful to Silvia Dávila-Manzanilla for her assistance with English grammar and vocabulary.
This work was supported by DGAPA-UNAM (IN 206212); by Fundación Miguel Aleman; and a PhD research fellowship awarded to Sitlali del Rosario Olguín-Reyes by CONACyT.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Heidelberger C. Studies on the cellular and molecular mechanisms of hydrocarbon carcinogenesis. Euro J Cancer 1970; 6: 161-172.
- Viau C, Hakizimana G, Bouchard M. Indoor exposure to polycyclic aromatic hydrocarbons and carbon monoxide in traditional houses in Burundi. Int Archiv Occupational Environ Health 2000; 73: 331-338.
- Perera FP, Hemminki K, Gryzbowska E, Motykiewicz G, Michalska J, Santella RM. Molecular and genetic damage in humans from environmental pollution in Poland. Nature 1992; 360: 256-258.
- Binkova B, Lewtas J, Miskova I, Lenicek J, Sram R. DNA adducts and personal air monitoring of carcinogenic polycyclic aromatic hydrocarbons in an environmentally exposed population. Carcinogenesis 1995; 16: 1037-1046.
- Perera F, Tang D, Whyatt R, Lederman SA, Jedrychowski W. DNA damage from polycyclic aromatic hydrocarbons measured by benzo (a) pyrene-DNA adducts in mothers and newborns from Northern Manhattan, the World Trade Center Area, Poland, and China. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the Am Assoc Cancer Res Am Soc Preventive Oncol 2005; 14: 709-714.
- Paget V, Lechevrel M, Andre V, Goff JL, Pottier D, Billet S. Benzo (a) pyrene, aflatoxine B (1) and acetaldehyde mutational patterns in TP53 gene using a functional assay: relevance to human cancer aetiology. PloS One 2012; 7: e30921.
- Yoshino I, Kometani T, Shoji F, Osoegawa A, Ohba T, Kouso H. Induction of epithelial-mesenchymal transition-related genes by benzo(a)pyrene in lung cancer cells. Cancer 2007; 110: 369-374.
- Hirano T, Franzen B, Kato H, Ebihara Y, Auer G. Genesis of squamous cell lung carcinoma. Sequential changes of proliferation, DNA ploidy, and p53 expression. Am J Pathol 1994; 144: 296-302.
- Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res 2008; 68: 9005-9014.
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006; 7: 21-33.
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990; 61: 759-767.
- Jimenez-Chillaron JC, Nijland MJ, Ascensao AA, Sardao VA, Magalhaes J, Hitchler MJ. Back to the future: transgenerational transmission of xenobiotic-induced epigenetic remodeling. Epigenetics 2015; 10: 259-273.
- Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016; 17: 284-299.
- Zhang G, Pradhan S. Mammalian epigenetic mechanisms. IUBMB Life 2014; 66: 240-256.
- Shimada T, Sugie A, Yamada T, Kawazoe H, Hashimoto M, Azuma E. Dose-response studies on the induction of liver cytochromes P4501A1 and 1B1 by polycyclic aromatic hydrocarbons in arylhydrocarbon-responsive C57BL/6J mice. Xenobiotica 2003; 33: 957-971.
- Brookes P. Mutagenicity of polycyclic aromatic hydrocarbons. Mutation Res 1977; 39: 257-283.
- Zhang Y, Dong S, Wang H, Tao S, Kiyama R. Biological impact of environmental polycyclic aromatic hydrocarbons (ePAHs) as endocrine disruptors. Environ Pollution 2016; 213: 809-824.
- Puga A, Marlowe J, Barnes S, Chang CY, Maier A, Tan Z. Role of the aryl hydrocarbon receptor in cell cycle regulation. Toxicology 2002; 182: 171-177.
- Volkov MS, Bolotina NA, Evteev VA, Koblyakov VA. Ah-receptor-independent stimulation of hepatoma 27 culture cell proliferation by polycyclic aromatic hydrocarbons. Biochem Biokhimiia 2012; 77: 201-207.
- Juan SH, Lee JL, Ho PY, Lee YH, Lee WS. Antiproliferative and antiangiogenic effects of 3-methylcholanthrene, an aryl-hydrocarbon receptor agonist, in human umbilical vascular endothelial cells. Euro J Pharmacol 2006; 530: 1-8.
- Teneng I, Montoya-Durango DE, Quertermous JL, Lacy ME, Ramos KS. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics 2011; 6: 355-367.
- Huang H, Hu G, Cai J, Xia B, Liu J, Li X. Role of poly(ADP-ribose) glycohydrolase silencing in DNA hypomethylation induced by benzo(a)pyrene. Biochem Biophys Res Communications 2014; 452: 708-714.
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 2007; 8: 286-298.
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010; 31: 27-36.
- Shen L, Fang J, Qiu D, Zhang T, Yang J, Chen S. Correlation between DNA methylation and pathological changes in human hepatocellular carcinoma. Hepato-Gastroenterology 1998; 45: 1753-1759.
- Tao L, Yang S, Xie M, Kramer PM, Pereira MA. Hypomethylation and overexpression of c-jun and c-myc protooncogenes and increased DNA methyltransferase activity in dichloroacetic and trichloroacetic acid-promoted mouse liver tumors. Cancer Lett 2000; 158: 185-193.
- Feng Y, Xue WJ, Li P, Sha ZY, Huang H, Rui L. RASSF1A hypermethylation is associated with aflatoxin B1 and polycyclic aromatic hydrocarbon exposure in hepatocellular carcinoma. Hepato-Gastroenterology 2012; 59: 1883-1888.
- Amenya HZ, Tohyama C, Ohsako S. Dioxin induces Ahr-dependent robust DNA demethylation of the Cyp1a1 promoter via Tdg in the mouse liver. Scientific Rep 2016; 6: 34989.
- Liu WB, Cui ZH, Ao L, Zhou ZY, Zhou YH, Yuan XY. Aberrant methylation accounts for cell adhesion-related gene silencing during 3-methylcholanthrene and diethylnitrosamine induced multistep rat lung carcinogenesis associated with overexpression of DNA methyltransferases 1 and 3a. Toxicol Appl Pharmacol 2011; 251: 70-78.
- Liu WB, Liu JY, Ao L, Zhou ZY, Zhou YH, Cui ZH. Dynamic changes in DNA methylation during multistep rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Toxicol Lett 2009; 189: 5-13.
- Rajendran P, Ho E, Williams DE, Dashwood RH. Dietary phytochemicals, HDAC inhibition, and DNA damage/repair defects in cancer cells. Clin Epigenet 2011; 3: 4.
- Adenuga D, Yao H, March TH, Seagrave J, Rahman I. Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. Am J Respir Cell Mol Biol 2009; 40: 464-473.
- Chang CC, Sue YM, Yang NJ, Lee YH, Juan SH. 3-Methylcholanthrene, an AhR agonist, caused cell-cycle arrest by histone deacetylation through a RhoA-dependent recruitment of HDAC1 and pRb2 to E2F1 complex. PloS One 2014; 9: e92793.
- Xie HJ, Noh JH, Kim JK, Jung KH, Eun JW, Bae HJ. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PloS One 2012; 7: e34265.
- Buurman R, Gurlevik E, Schaffer V, Eilers M, Sandbothe M, Kreipe H. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology 2012; 143: 811-820.
- Quint K, Agaimy A, Di Fazio P, Montalbano R, Steindorf C, Jung R. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Archiv 2011; 459: 129-139.
- Lachenmayer A, Toffanin S, Cabellos L, Alsinet C, Hoshida Y, Villanueva A. Combination therapy for hepatocellular carcinoma: additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J Hepatol 2012; 56: 1343-1350.
- Fan J, Lou B, Chen W, Zhang J, Lin S, Lv FF. Down-regulation of HDAC5 inhibits growth of human hepatocellular carcinoma by induction of apoptosis and cell cycle arrest. Tumour Biol J Int Soc Oncodevelopmental Biol Med 2014; 35: 11523-11532.
- Feng GW, Dong LD, Shang WJ, Pang XL, Li JF, Liu L. HDAC5 promotes cell proliferation in human hepatocellular carcinoma by up-regulating Six1 expression. Euro Rev Med Pharm Sci 2014; 18: 811-816.
- Lv Z, Weng X, Du C, Zhang C, Xiao H, Cai X. Downregulation of HDAC6 promotes angiogenesis in hepatocellular carcinoma cells and predicts poor prognosis in liver transplantation patients. Mol Carcinogenesis 2016; 55: 1024-1033.
- Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harbor Perspectives Med 2016; 6.
- Gnyszka A, Jastrzebski Z, Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res 2013; 33: 2989-2996.
- Gallagher S, Winston SE, Fuller SA, Hurrell JG. Immunoblotting and immunodetection. Curr Protocols Immunol 2008.
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671-675.
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Communications 2013; 4: 1563.
- Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure 2008; 16: 341-350.
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. New England J Med 2003; 349: 2042-2054.
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004; 429: 457-463.
- Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Ann Rev Pharmacol Toxicol 2005; 45: 629-656.
- Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res 2006; 66: 8342-8346.
- Stellas D, Souliotis VL, Bekyrou M, Smirlis D, Kirsch-Volders M, Degrassi F. Benzo(a)pyrene-induced cell cycle arrest in HepG2 cells is associated with delayed induction of mitotic instability. Mutation Res 2014; 769: 59-68.
- Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem 2005; 280: 18302-18310.
- Sun L, Hui AM, Kanai Y, Sakamoto M, Hirohashi S. Increased DNA methyltransferase expression is associated with an early stage of human hepatocarcinogenesis. Japanese J Cancer Res Gann 1997; 88: 1165-1170.
- Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol 2004; 164: 689-699.
- Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology 2001; 33: 561-568.
- Xia B, Yang LQ, Huang HY, Pang L, Yang XF, Yi YJ. Repression of biotin-related proteins by benzo(a)pyrene-induced epigenetic modifications in human bronchial epithelial cells. Int J Toxicol 2016; 35: 336-343.
- Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene 2005; 24: 13-19.
- Huang HY, Cai JF, Liu QC, Hu GH, Xia B, Mao JY. Role of poly (ADP-ribose) glycohydrolase in the regulation of cell fate in response to benzo(a)pyrene. Experimental Cell Res 2012; 318: 682-690.
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002; 21: 5483-5495.
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009; 10: 32-42.
- Parbin S, Kar S, Shilpi A, Sengupta D, Deb M, Rath SK. Histone deacetylases: a saga of perturbed acetylation homeostasis in cancer. J Histochem Cytochem 2014; 62: 11-33.
- Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007; 26: 5420-5432.
- Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. The Prostate 2004; 59: 177-189.
- Huang BH, Laban M, Leung CH, Lee L, Lee CK, Salto-Tellez M. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differentiation 2005; 12: 395-404.
- Song J, Noh JH, Lee JH, Eun JW, Ahn YM, Kim SY. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS: Acta Pathologica, Microbiologica, Et Immunologica Scandinavica 2005; 113: 264-268.
- Hrzenjak A, Moinfar F, Kremser ML, Strohmeier B, Staber PB, Zatloukal K. Valproate inhibition of histone deacetylase 2 affects differentiation and decreases proliferation of endometrial stromal sarcoma cells. Mol Cancer Therap 2006; 5: 2203-2210.
- Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem 2006; 281: 13548-13558.
- Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol 2003; 160: 1017-1027.
- Basile V, Mantovani R, Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J Biol Chem 2006; 281: 2347-2357.
- Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics 2014; 6: 139-150.
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci 2009; 5: 147-152.
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001; 107: 137-148.
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001; 107: 149-159.
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 2005; 123: 437-448.