- Biomedical Research (2009) Volume 20, Issue 1
Structural interaction between drug - DNA and protein- A novel approach for bioinformatics in medicine
Ajit. K Saxena*, Divya Singh and Gajendra SinghHuman Cytogenetic Laboratory, Centre of Experimental Medicine and Surgery, Institute of Medical Sciences, Banaras Hindu University, Varanasi, India
- *Corresponding Author:
- Ajit K Saxena
Human Cytogenetics Laboratory,
Centre of Experimental Medicine and Surgery,
Institute of Medical Sciences
Banaras Hindu University
Varanasi 221005
India
E-mail: draksaxena1@rediffmail.com
Accepted date: October 26 2008
Abstract
Drug like methotrexate were used in the present study because of dual impor-tance, both for clinicians as well as an interest of basic scientists. Methotrexate, an antineoplastic agent is used for malignant as well as non malignant conditions and become teratogenic & mutagenic in nature. They prevent cell proliferation by incorporated into DNA during the "S"- phase of the cell- cycle and inhibit rapidly dividing normal somatic cell-division including germ cells because of an-tagonist of folic acid synthesis by inhibiting dihydrofolate reductase an essential enzyme required for the conversion of folic acid to tetrahydrofolic acid. The ra-tional behind this study is to observe binding sites activity of native methotrexate structure to DNA, and dihydrofolate reductase (protein) as well as mutated structure of drug (methotrexate) using different software of structural modeling. For drug (methotrexate) - protein structure interaction, homology modeling, also known as comparative modeling, was used in the present study for con-structing model of a protein from its amino acid sequence to enhance activity of the drug by structural modification. All homology modeling techniques help for the identification of one or more known protein structures (known as "tem-plates" or "parent structures") resemble to the structure of query sequence, and on the production of an alignment, which maps residues in the query sequence. The sequence alignment and template structure are then used to produce a structural model of the target site of drug or protein. Interestingly in the present study indicates the significant interaction between the drug and DNA/ proteins were noticed which could be further useful in reaching qualitative and quantita-tive conclusions about the query sequence of developing new molecule (drug ac-tion), especially in formulating hypotheses about why certain residues are con-served and making toxic side effects which may in turn lead to experiments to test hypotheses of drug mediated toxicity.Keywords
Methotrexate, DNA, Dihydrofolate reductase
Introduction
Drug interaction is a reaction in which a substance affects the activity of a drug, i.e. the effects are increased or de-creased, or to produce new effect(s) that neither produces on its own, depend upon the active sites in a molecule. Drug protein binding is the reversible interaction of drugs with proteins in plasma. Proteins are large organic com-pounds consists of different kinds of amino acids ar-ranged in a linear form and joined together by peptide bonds between the carboxyl and amino groups of adjacent amino acid residues. The residual proteins are sometime chemically altered in post-translational modification: ei-ther before the protein can function in the cell, or as part of control mechanisms. Proteins can also work together to achieve a particular function, and they often associate to form stable complexes. Like other biological macromole-cules such as polysaccharides and nucleic acids, proteins are essential parts of organisms and participate in every process within cells. Many proteins are enzymatic in na-ture that catalyze biochemical reactions and are vital to metabolism. DNA is often compared to a set of blueprints or a recipe, since it contains the instructions needed to construct other components of cells, such as proteins and RNA molecules [1].
Many mutagens including methotrexate fit into the space between two adjacent base pairs, this is called intercalat-ing. Most intercalators are aromatic and planar molecules, and include ethidium, daunomycin, doxorubicin and tha-lidomide. In order for an intercalator to fit between base pairs, the bases must separate, distorting the DNA strands by unwinding of the double helix. This inhibits both tran-scription and DNA replication, causing toxicity and muta-tions. As a result, DNA intercalators are often carcino-gens, and other molecules like benzopyrene diol epoxide, acridnes, aflatoxin and ethidium bromide are also well-known examples. Nevertheless, due to their ability to in-hibit DNA transcription and replication, these toxins are also used in chemotherapy to inhibit rapidly-growing can-cer cells [2].
The binding of drug to protein is a saturable process gov-erned by the same mass action expression that describes the interaction of a substrate with an enzyme binding site. The extent to which a drug is bound in plasma or blood is usually expressed as the fraction unbound. The fraction unbound to a drug is determined by 1) the affinity of the drug for the protein 2) the concentration of the binding protein, and 3) the concentration of drug relative to that of the binding protein [3].
There are three principally different ways of drug-binding. First, through control of transcription factors and polymerases, where, the drugs interact with the proteins that bind to DNA. Second, through RNA binding to DNA double helices to form nucleic acid triple helical struc-tures or RNA hybridization (sequence specific binding) to exposed DNA single strand regions forming DNA-RNA hybrids that may interfere with transcriptional activity. Third, small aromatic ligand molecules that bind to DNA double helical structures by (i) intercalating between stacked base pairs thereby distorting the DNA backbone conformation and interfering with DNA-protein interac-tion or (ii) the minor groove binders. The lack of se-quence specificity for intercalating molecules, however, does not allow to target specific genes, but rather certain cellular states or physiological and pathological condi-tions. A detailed enumeration and quantification of these energetic contributions can help in addressing vital issues involved in predicting ligand binding free energies and developing a viable methodology for designing or modi-fying DNA binding small molecules with structural and chemical features. This would allow them to recognize the properties of DNA sequences which bind to them with requisite sequence specificity and binding affinity, such that a competition with regulatory proteins could ensue. Developing a molecular view of the thermodynamics of DNA recognition is essential to the design of ligands for regulating gene expression. A novel energy component strategy makes it possible to study the thermodynamics of DNA-drug binding make correlations between structure and energetic [4].
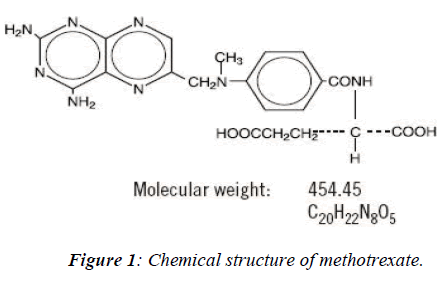
Methotrexate, an anti-neoplastic agent developed for the treatment of cancer in 1988 the US Food and Drug Ad-ministration (FDA) approved also for the treatment of psoriasis. The IUPAC name of methotrexate is 4-amino-4-deoxy-10-methyl-pteroyl-glutamic acid. Its chemical formula is C20H22N8 O5 and molecular weight is 454.45 belongs to the class of antimetabolites. This drug a folate antagonist is employed in the treatment of various kinds of neoplasm as well as psoriasis and also being used in combination with other chemotheraptic agents for the treatment of acute lymphoblastic leukemia or myeloblas-tic leukemia. Folic acid, an essential component required for DNA synthesis through folate metabolism which re-quires a co-enzyme- tetrahydrofolate reductase converts tetrafolic acid into dihydrofolic acid. Methotrexate is highly toxic in nature because it entered into the S-phase of the cell-cycle of rapidly dividing cells such as in fetus cells, germ cells, liver and bone marrow [5,6,7], leads to inhibit DNA replication and finally cell-death. The main aim is of the present study is how to reduce the terato-genicity or mutagenicity or drug mediated cell toxicity /damage by structural remodeling and their binding sites modification with protein or an intermediate enzyme (DHFR) using different softwares [8].
Materials and Methods
Drug-like methotrexate is selected because of dual impor-tance to the interest of clinicians as well as to the basic scientists. The binding activity of methotrexate drugs with other molecules including DNA & protein (enzyme)were studied by using different softwares of bioinformatics tools like SPDB Viewer, Rasmol, EMBOSS win and also use of freely available tools like CAST p, EXPASY to determine secondary structure and prediction of structure in the present study.
a) Swiss PDB Viewer
Swiss-Pdb Viewer is an application that provides a user friendly interface allowing analyzing several proteins at the same time. The proteins can be superimposed in order to deduce structural alignments and compare their active sites or any other relevant parts. Amino acid mutations, H-bonds, angles and distances between atoms are easy to obtain for the spontaneous graphic and menu interface.
This is an automated homology modeling server devel-oped within the Swiss Institute of Bioinformatics (SIB) in collaboration between GlaxoSmithKline R&D and the Structural Bioinformatics Group at the Biozentrum in Basel. These two programs reduces the amount of work necessary to generate models, as it is possible to thread a protein primary sequence onto a 3D template and get an immediate feedback of how well the threaded protein will be accepted by the reference structure before submitting a request to build missing loops and refine side chain pack-ing. Swiss-PdbViewer can also read electron density maps, and provides various tools to build into the density. In addition, various modeling tools are integrated and command files for popular energy minimization packages can be generated.
b) Rasmol
Rasmol is another computer program written for molecu-lar graphics visualization intended and used primarily for the depiction and exploration of biological macromole-cule structures, such as those found in the Protein Data Bank. It was originally developed by Roger Sayle in the early 90s. Rasmol has become an important educational tool as well as continuing to be an important tool for re-search in structural biology. Rasmol has a complex ver- sion starting with the series of 2.7 versions.
c) Emboss
EMBOSS is the commercial "European molecular biol-ogy open software suite". EMBOSS is a free Open Source software analysis package specially developed for the needs of the molecular biology (e.g.EMBnet) user com-munity. This software automatically copes with data in a variety of formats and even allows transparent retrieval of sequence data. It also integrates a range of currently available packages and tools for sequence analysis.
Results and Discussion
Bioinformatics involves the manipulation, searching, and data mining of DNA sequence data. The development of techniques to store and search DNA sequences have led to widely-applied advances in computer science, especially string searching algorithms, machine learning and data-base theory. String searching or matching algorithms, which find an occurrence of a sequence of letters inside a larger sequence of letters, were developed to search for specific sequences of nucleotides (9). In other applications such as text editors, even simple algorithms for this problem usually suffice, but DNA sequences cause these algorithms to exhibit near-worst-case behaviour due to their small number of distinct characters. The problem related to sequence alignment aims to identify homolo-gous sequences and locate the specific mutations that make them distinct. These techniques, especially multiple sequence alignment, are used in studying phylogenetic relationships and protein function.
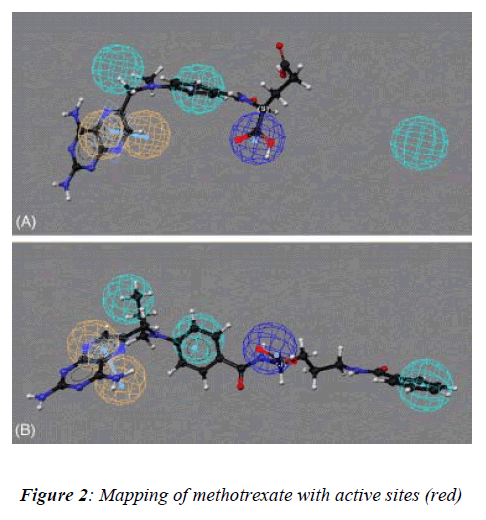
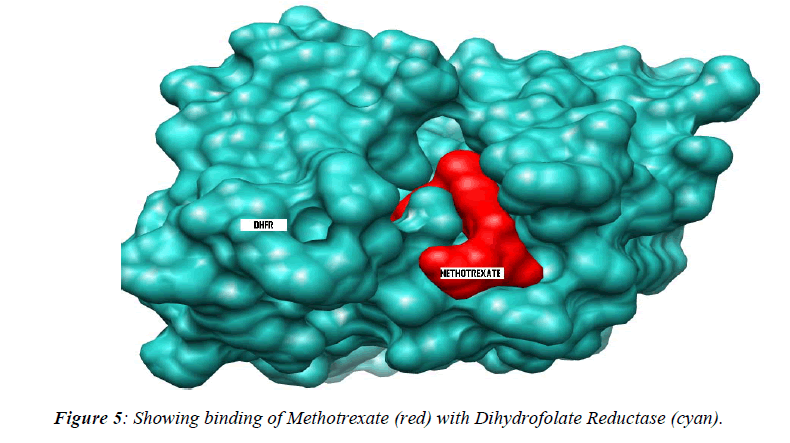
In our present study, the 3D structure reveals binding sites of the methotrexate (red in appearance) as shown (Figure-2) with help of the softwares as describe above to under-stand the number of chains, groups, bonding, helices and turns were observed during analysis of data of meth-otrexate structure which have a tendency to interact with other molecules (drug) covalently [10]. These softwares have several advantages: a) properly constructed toolkit for creating robust bioinformatics applications or work-flows, b) comprehensive set of sequence analysis programs, c) all sequence with many alignment and structural formats are handled, d) extensive programming library for sequence analysis tasks, e) additional programming librar-ies for many other areas including string handling, patt-ern-matching, list processing and database indexing,
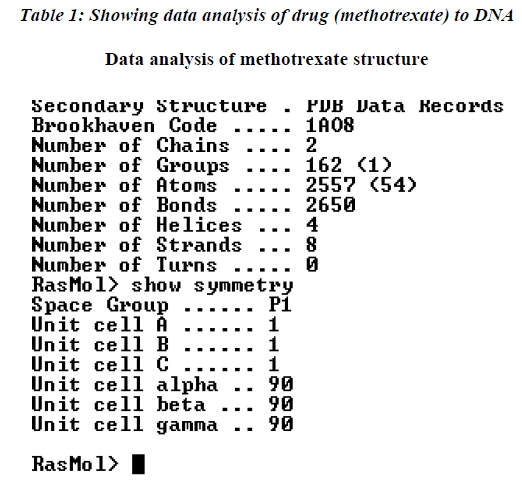
The further analysis of the drug binding was carried out to the DNA molecule showing the interaction with number of amino acids as shown in Table 1. The data was further analyzed which indicate that maximum affinity of drug to DNA was with leucine while minimum was recorded with cysteine amino acid residue.
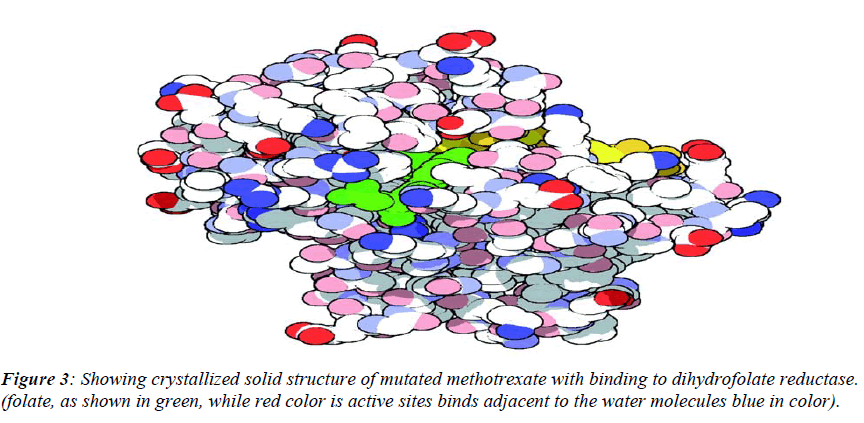
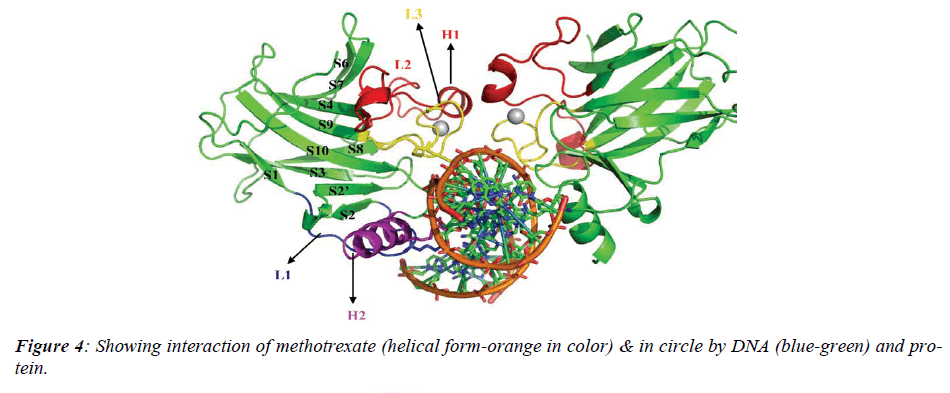
The native structure of the drug was mutated with the help of above software to enhance the active sites with efficacy of the drug to bind with DNA and protein mole-cule as shown in crystalline form of structure (Figure.3), which revealed that maximum number of affinity towards the amino acid (leucine) enhance in mutant structure but the minimum amino acids which bind to DNA was his-tidine instead of cysteine.The above table showing meth-otrexate binding sites with DNA (Figure. 4) using online software CASTp. The structure has been developed to observe that how to increase the active site. Therefore we have developed the mutated structure of the original drug (methotrexate) to enhance the efficiency of the drug by using different bioinformatics tools i.e. softwares. Table 1 is showing data of normal methotrexate with DNA in which the maximum number of amino acid was found to be 18 which is leucine and a minimum of one amino acid which is cysteine.
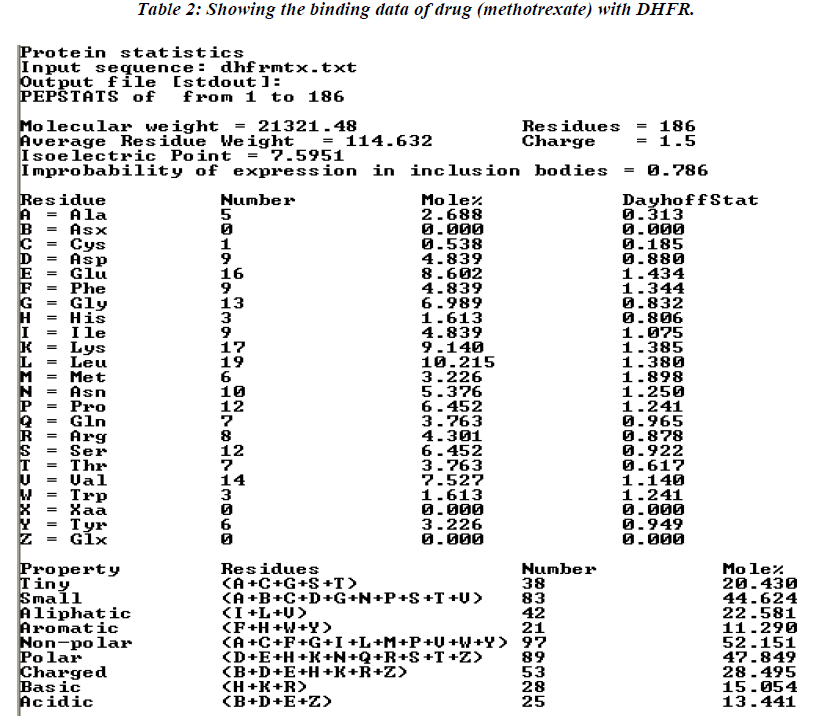
The thermodynamics and kinetics of the interaction of dihydrofolate reductase (DHFR) with methotrexate have been studied by using fluorescence, stopped-flow, and single-molecule methods. DHFR was modified to permit the covalent addition of a fluorescent molecule [11]. Ta-ble 2 showing data analysis of normal methotrexate with dihydrofolatereductase. Further analysis of the present study reveals that maximum number (17) of amino acid residue affinity with lysine while minimum with trypto-phan residue as details were presented in tabulated form. The present study was documented in tabulated form which showing that mutated methotrexate with DNA re-veals the increasing number of binding sites with amino acid residues i.e. 19 leucine. While minimum amino acid binding residue is replaced by histidine instead of cys-teine residue , although the authors are unable to conclude the replacement of such amino acid residue but it believe that might have associated with activity of the drug. However, the study should be further continued to con-firm the activity of the mutated drug using different bioin-formatics softwares.
Conclusions
Although, to enhance the activity of drug it is necessary to modify the native structure of the drug by either increas-ing or decreasing active sites. The basic parameter that is used in torsion angles, which depends on four atoms. The system taken is two linked peptide units. After deciding the above things parameter is decided. The two bonds N-Cα and Cα-N are the one around which rotation are per-mitted and hence the torsion angles have been defined with respect to these bonds.In human methotrexate is given to the malignant as well as non-malignant patients. It is an interesting molecule (MTX) to identify drug -protein interaction with or with out DNA to identify the nature of active sites responsible for binding activity with these molecules, left behind unanswerable certain ques-tions like how to reduce the mutagenicity of the drug as-sociated with birth defects? Therefore the present study has been designed with aim to develop the mutant struc-ture of the drug and shows enhance binding activity due to increase active sites. Similarly, these findings are fur-ther confirmed by drawing Ramachandran plot and amino acid analysis using different soft wares showing significant results. These preliminary short term studies have immense value for future designing and developing new molecule (drug) and their pharmacokinetics study.
Acknowledgement
AKS is thankful to Ms Archna Singh and Mr. M. Murti for providing technical assistance during the preparation and data analysis of the manuscript.
References
- Lindahl T. Instability and decay of the primary struc- ture of DNA. Nature 1993, 362: 709-715.
- Controlling occupational exposure to hazardous drugs (OSHA Work-Practice Guidelines). J Health Syst Pharm 1996; 53: 1669-1685
- Chen MJ, Shimada T, Moulton AD, Cline A, Humph- ries RK, Maizel J, Nienhuis AW. The functional human dihydrofolate reductase gene. J Biol Chem. 1984; 259: 3933-3943.
- eFriedberg EC. DNA damage and repair. Nature 2003, 421: 436-440.
- Jordan RL, Wilson JG, Schumacher HJ. Embryo toxic- ity of folate antagonist methotrexate in rat and rabbits. Teratology. 1977; 15: 73-79.
- Saxena A K, Dhungel S, Bhattacharya S, Srivastava A K. Effect of chronic low dose of methotrexate on cellu- lar proliferation during spermatogenesis in rat. Arch Andol. 2004; 50: 33-35.
- West SG. Methotrexate hepatotoxicity. Rheum Dis Clin North Am. 1997; 23: 883-915
- Thomas LL, Mertens MJ, von dem Borne AE, van Boxtel CJ, Veenhof CH, Veies EP. Clinical manage- ment of cytotoxic drug overdose. Med Toxicol Adverse Drug Exp 1988; 3: 253-263
- Masters JN, Attardi G. The nucleotide sequence of the c DNA coding for the human dihydrofolate reductase. Gene .1983; 21: 59-63.
- Davies JF, Delcamp TJ, Prendergast NJ, Ashford VA, Freisheim JH, J.Kraut J. Crystal structures of recombi- nant human Dihydrofolate Reductase complexed with folate and Methotrexate. Nature 1990; 29: 779-784.
- Davies JF II, Delcamp TJ, Prendergast NJ, Ashford VA, Freisheim JH, Kraut J. Crystal structures of reco- mbinant human dihydrofolate reductase complexed with folate and 5-deazafolate. Biochemistry. 1990; 9: 29-40.