Research Article - Biomedical Research (2017) Volume 28, Issue 9
Stability of bovine serum albumin labelled by rhodamine B isothiocyanate
Ting Yang#, Dali Sun#, Pengyuan Xu*, Shumin Li, Yunyun Cen, Yijun Li, Qinwen Xu, Yanbo Sun,Weiming Li, Yueying Lin, Yuxing Qi and Xiongzhi ChenDepartment of Medical Cares for Cadres, the Second Affiliated Hospital of Kunming Medical University, Kunming, Yun Nan Province, China
#These authors contributed equally to this work
- *Corresponding Author:
- Pengyuan Xu
Department of Medical Cares for Cadres
Surgical nutritional research center of Yun Nan Province, China
Accepted on January 25, 2017
Abstract
The aim of this study was to label Bovine Serum Albumin (BSA) by Rhodamine B Isothiocyanate (RBITC) and test its stability in vivo. RBITC-BSA was labelled by the improved Marshall’s method, scanned at full wavelength, and the RBITC:BSA ratio was calculated. Blood and urine samples were obtained from rabbits injected with the RBITC-BSA complex and SDS-PAGE analysis was performed. Full wavelength scanning showed that the labelled RBITC-BSA complex possessed features of both RBITC and BSA, and the RBITC:BSA ratio was 1:80. Furthermore, the RBITC-BSA complex was stable in vivo. Thus, we developed a stable RBITC-BSA complex with high specificity and sensitivity, which could be used as a tracer molecule to study protein transportation and vascular permeability.
Keywords
Rhodamine B isothiocyanate, Bovine serum albumin, Full wavelength scanner, SDS-PAGE.
Introduction
With the recent development of fluorescence-based analytical approaches in medicine and biochemistry, several fluorescent probes are being used in biological analysis. Presently, fluorescent probes include small molecules [1-4], nanoprobes [5-7], luminescent lanthanide complexes [8-10], and certain fluorescent proteins [11-14]. Protein-based fluorescent probes are used to detect physiological activities of proteins, which is important for clinical treatment. Since the first report of a fluorescent protein antibody in 1950 by Coons and Kaplan [15], there has been an increase in the development and use of fluorescent small molecule-protein combinations in bioanalysis, which has played pivotal roles in biomedical and basic research.
Fluorescent isothiocyanate is a common fluorescent probe, which has high quantum yield, superior light stability, and low temperature coefficient. Under alkaline conditions, the isothiocyano group of the fluorescent isothiocyanate could directly combine with the amino moiety of proteins via a phosphoryl reaction to form a fluorescein-protein complex. Fluorescein-albumin is as fluorescent tracer molecule, which is used to investigate vascular permeability [16]. A fluoresceinalbumin complex developed by a statistically significant method must possess the following characteristics: 1) the fluorescein must be conjugated to the protein in vitro; 2) the fluorescein-albumin complex should be separable from the non-reacted fluorescein and its degradation products [17].
However, studies reporting the stability of the RBITC-BSA complex as a fluorescent tracer in vivo are lacking. To investigate whether the RBITC-BSA complex was suitable as a tracer molecular in vivo, we designed an RBITC-BSA complex; the RBITC was removed by glucose gel chromatography and analysed by full wavelength scanning. Moreover, serum and urine samples from rabbits at different time points were analysed to determine the stability of the RBITC-BSA by Sodium Dodecyl-Polyacrylamide Gel Electrophoresis (SDS-PAGE).
Materials and Methods
Preparation of the RBITC-BSA complex
BSA was labelled by the improved Marshall’s method. Saline (0.15 M, NaCl) and a buffer solution (0.15 M, NaHCO3/ Na2CO3, pH 9.0) were mixed in a 9:1 (v/v) ratio to form a 10 mg/ml protein solution. Then, RBITC, in which the M ratio of protein to fluorescein was 5:1, was added slowly into the protein solution, followed by stirring for 12 h in the dark at 4°C. After that, the mixed solution was placed in a filter bag and dialyzed against phosphate buffer saline (0.01 M PBS, pH 7.4) at 4°C in the dark for 2 days.
A Sephadex G-75 column (Superfine, GE Healthcare, 2.6 cm diameter, 100 cm length) was equilibrated with phosphate buffer (8 g/L NaCl, 0.2 g/L KCl, 0.2 g/L KH2PO4•12H2O, pH 7.4), followed by loading of specimens at 4°C, and dialysis in the dark. The column was washed with PBS at a speed of 10 min/3 ml (constant flow pump BT100-1F, Baoding Lange Constant Flow Pump Co., Ltd) and 3 ml eluates were collected per tube (automatic collector BS-100A, Shanghai Qingpuhuxi instrument factory). The eluate corresponding to the first fluorescent band from the column was observed and recorded. After collecting approximately 500 ml of eluate, each tube was examined by an Ultraviolet (UV) spectrophotometer (Ultrospec 2100 pro, GE) and absorbance values at 280 nm were calculated.
Confirmation of the light absorption characteristics
In the RBITC-BSA complex, the RBITC and BSA were subjected to full wavelength scanning (190-900 nm) by the UV-visible spectrophotometer. The changes in the light absorption characteristics of RBITC-BSA, RBITC, and BSA were compared.
Detection of labelling rate
The protein (λ=230 nm) and fluorescein levels (Ex: 555 nm, Em: 582 nm) in the eluting peak were detected by the UVvisible spectrophotometer and quantitated by the standard curve method. The formula to calculate the molecular ratio of RBITC and BSA in the product was F/P=(fluorescein level (μg/ml) × 10-3/fluorescein molecular weight)/(protein level (mg/ml)/albumin molecular weight).
Preparation of specimens in vivo
Three milliliters of the RBITC-BSA complex was injected in healthy male Japanese rabbits weighing 2.5 kg. Thereafter, 2 ml blood from the right ear vein (experimental) of the rabbits was collected at 10 min and 30 min after the injection, respectively. Similar amount of blood was drawn from the left ear vein as a control before the injection. Blood specimens were marked as 0, 10, and 30 that corresponded to 0 min, 10 min, and 30 min samples, respectively. Two milliliters urine was obtained from bladder.
SDS-PAGE analysis
Five microliters specimen (about 30-50 μg protein) was used for SDS-PAGE on a 12% gel at 120 V for 80 min. First, the fluorescent signal on the gel was imaged, followed by staining with Coomassie Brilliant Blue R250 for 1 h. After that, the gel was destained overnight. Finally, the gel was photographed by an electrophoretic image analyzer (Bio-RAD Universal HoodII)
Results
Chromatography
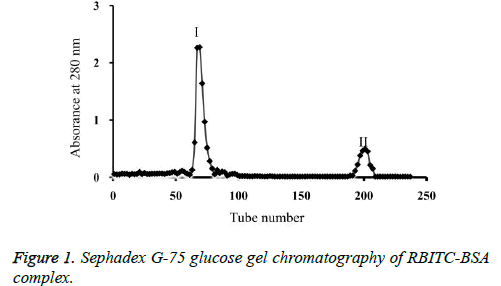
The 60th eluate tube of the Sephadex G-75 column chromatography of the RBITC-BSA complex corresponded to the appearance of the first fluorescent band. There were two eluting peaks corresponding to the Optical Density 280 (OD280) values (Figure 1); the first peak occurred in fractions 60-80 (eluting peak I) and the second peak in fractions 180-220 (eluting peak II). Based on the principle of glucose gel chromatography, the eluting peak I corresponded to the RBITC-labelled BSA, which was collected. The eluting peak II was free RBITC, which was discarded.
The OD280 readings of the eluate fractions showed two values in the UV-visible spectrophotometer; the first value corresponded to tube numbers 60-80 (eluting peak I), which contained the RBITC-labelled BSA; these fractions were combined; the second value corresponded to tube numbers 180-220 (eluting peak II), which was free RBITC, and were discarded.
Verification of the light absorption characteristics of RBITC
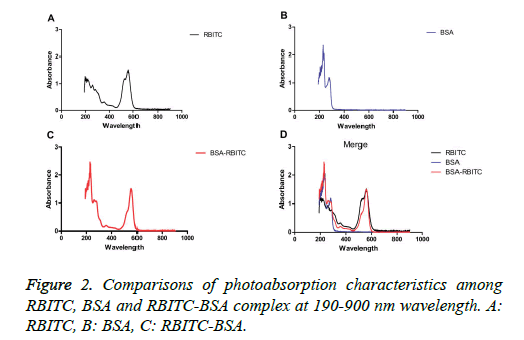
The maximal (at 555 nm) and minimal photoabsorption (at 197 nm) of RBITC (Figure 2A) and the maximum photoabsorption of BSA at 555 nm (Figure 2B) were assessed by full wavelength scanning. However, the eluting peak I had two maxima of photoabsorption at 230 nm and 558 nm, respectively (Figure 2C), which indicated that the eluting peak was the RBITC-labelled BSA complex.
RBITC had the maximal light absorption at 555 nm and minimal light absorption at 197 nm (Figure 2A), whereas BSA had the maximal light absorption at 230 nm (Figure 2B). The fractions containing the eluting peak I (Figure 1) had two maxima of light absorption at 230 nm and 558 nm, thereby confirming the presence of the RBITC-BSA complex (Figure 2C).
Calculation of the marked rate
The F/P value of fractions 60-80 (first eluting peak) of the Sephadex G-75 gel chromatography was 1.80. The F/P value reflects the amount of protein marked by fluorescein. Generally, the value of F/P ranges from 1 to 2. A high value indicates non-specific marking, whereas low value indicates weak fluorescence and low sensitivity.
SDS-PAGE
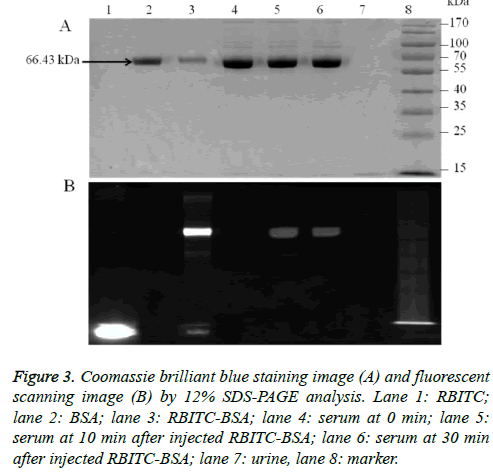
SDS-PAGE analysis of the serum obtained from rabbits injected with the RBITC-BSA complex showed fluorescent signal in the 10 min and 30 min post-injection samples; the position of fluorescent band was identical to the position of the chromatographically purified RBITC-BSA (Figure 3B). This result suggested that the RBITC-BSA complex was stable before and after the in vivo experiment. Coomassie Brilliant Blue R250 staining showed that the purified RBITC-BSA complex and the fluorescent bands observed in the 10 min and 30 min post-injection serum samples electrophoresed at 66.43 kDa, which is the size of BSA. This indicated that BSA was also stable in the serum; however, the protein was not observed in the urine samples under identical conditions (Figure 3A).
Figure 3: Coomassie brilliant blue staining image (A) and fluorescent scanning image (B) by 12% SDS-PAGE analysis. Lane 1: RBITC; lane 2: BSA; lane 3: RBITC-BSA; lane 4: serum at 0 min; lane 5: serum at 10 min after injected RBITC-BSA; lane 6: serum at 30 min after injected RBITC-BSA; lane 7: urine, lane 8: marker.
Discussion
Fluorescent isothiocyanate is a probe with superior fluorescence characteristics. The isothiocyano group of fluorescent isothiocyanate can directly conjugate with the amino group of proteins to form sulfur-hydrocarbon bonds via a phosphoryl reaction to yield a fluorescently labelled protein probe. The isothiocyano group of RBITC can bind to the amino group of proteins (mainly ε-NH of lysine) under alkaline conditions, and yield a fluorescent protein probe via the formation of a carbon-sulfur-amino bond, which involves acylation of carbon [18]. Although the tertiary structure of proteins changed at pH 9.0 during the formation of the RBITCBSA complex, the complex still retained its secondary helical structure [19]. Andersson et al. [20] found three binding sites for fluorescein in BSA, which utilize both stable covalent and unstable non- covalent bonds. BSA could combine with RBITC via stable covalent bonds after the scrambling of its three-dimensional structure in the alkaline pH of the reaction. The non-covalent bonding between RBITC and BSA was disrupted by chloride ions of the buffer solution used in glucose gel chromatography. Full wavelength scanning indicated that the first peak obtained from the Sephadex column contained the BSA-RBITC complex, which was formed via covalent bonding. This complex was injected into rabbits; SDS-PAGE analysis of the post-injection serum samples revealed the presence of fluorescently labelled proteins, which were of the same size as that of BSA. This indicated that the RBITC-BSA complex was stabile in vivo and could be used as a tracer in studies regarding capillary permeability and molecule transfer.
Conclusion
We successfully prepared a RBITC-BSA complex, which is highly specific, sensitive, and stable in vitro, and could be used as a tracer molecule for study of protein transportation and vascular permeability.
Acknowledgements
Project supported by the National Natural Science Foundation of China (Grant No.81160114) and the Special foundation of Yun Nan province (Grant NO.2011FB196).
Conflicts of Interest
All of the authors declare that they have no conflicts of interest regarding this paper.
References
- Terai T, Nagano T. Small-molecule fluorophores and fluorescent probes for bioimaging. Pflügers Arch Eur J Physiol 2013; 465: 347-359.
- Chan J, Dodani SC, Chang CJ. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nature Chem 2012; 4: 973-984.
- Hao Z, Song X, Zhu M, Meng X, Zhao S, Su S, Yang W, Song S, Zhang H. One-dimensional channel-structured Eu-MOF for sensing small organic molecules and Cu2+ ion. J Mater Chem A 2013; 1: 11043-11050.
- Wagner BK, Carrinski HA, Ahn YH, Kim YK, Gilbert TJ, Fomina DA, Schreiber SL, Chang YT, Clemons PA. Small-molecule fluorophores to detect cell-state switching in the context of high-throughput screening. J Am Chem Soc 2008; 130: 4208-4209.
- Chen X, Li L, Xie L. Preparation and characterization of fluorescent chitosan nanoprobe. J Third Milit Med Univ 2013.
- Pellach M, Goldshtein J, Ziv-Polat O, Margel S. Functionalised, photostable, fluorescent polystyrene nanoparticles of narrow size-distribution. J Photochem Photobiol A Chemistry 2012; 228: 60-67.
- Huber V, Sereda G, Dale J. Rapid and inexpensive assay for evaluation of antibody efficacy with custom-designed fluorescent nanoparticles. US20140287438 2014.
- Terai T, Ito H, Kikuchi K, Nagano T. Salicylic-acid derivatives as antennae for ratiometric luminescent probes based on lanthanide complexes. Chem A Eur J 2012; 18: 7377-7381.
- Brooks R, Du Z, Borlace G, Brooks D, PlushS. Synthesis and characterisation of first generation luminescent lanthanide complexes suitable for being adapted for uptake via the mannose receptor. J Inorg Chem 2013; 2013: 1-8.
- Andrews M, Jones JE, Harding LP, Pope SJ. Luminescent probes based on water-soluble, dual-emissive lanthanide complexes: metal ion-induced modulation of near-IR emission. Chem Commun 2010; 47: 206-208.
- Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science 2011; 333: 642-646.
- Glick BS. Monomeric Red Fluorescent Proteins. J Biol 2011.
- Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol 2011; 29: 757-761.
- Dennis AM, Rhee WJ, Sotto D, Dublin SN, Bao G. Quantum dot-fluorescent protein FRET probes for sensing intracellular pH. ACS Nano 2012; 6: 2917-2924.
- Coons AH, Kaplan MH. Localization of antigen in tissue cells; improvements in a method for the detection of antigen by means of fluorescent antibody. J Exp Med 1950; 91: 1-13.
- Huang D, Wang L, Dong Y, Pan X, Li G, Wu C. A novel technology using transscleral ultrasound to deliver protein loaded nanoparticles. Eur J Pharm Biopharm 2014; 88: 104-115.
- Landel AM. Stability studies on fluorescein isothiocyanate--bovine serum albumin conjugate. Anal Biochem 1976; 73: 280-289.
- Weiss HR. Measurement of cerebral capillary perfusion with a fluorescent label. Microvasc Res 1988; 36: 172-180.
- Jr LW, Vijai KK, Foster JF. A structural transformation in bovine and human plasma albumins in alkaline solution as revealed by rotatory dispersion studies. J Biol Chem 1963; 238: 1984-1988.
- Andersson LO, Rehnstrom A, Eaker DL. Studies on nonspecific binding. Eur J Biochem 1971; 20: 371-380.