Short Communication - Biomedical Research (2019) Volume 30, Issue 2
Single step for Neisseria gonorrhoeae genomic DNA detection by using gold nanoparticle probe.
Paweena Tunakhun1, Pornsuda Maraming1, Ratree Tavichakorntrakool1, Phitsamai Saisud1, Saksit Sungkiri1, Sakda Daduang2, Patcharee Boonsiri3 and Jureerut Daduanga1,4*
1Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences, Khon Kaen University, Khon Kaen, Thailand
2Faculty of Pharmaceutical Sciences, Khon Kaen University, Thailand
3Department of Biochemistry, Faculty of Medicine, Khon Kaen University, Thailand
4HPV & EBV and Carcinogenesis Research Group, Khon Kaen University, Thailand
- *Corresponding Author:
- Dr. Jureerut Daduang
Centre for Research and Development of Medical Diagnostic Laboratories
Faculty of Associated Medical Sciences
Khon Kaen University, Thailand
Accepted date: March 21, 2019
DOI: 10.35841/biomedicalresearch.30-19-133
Visit for more related articles at Biomedical ResearchAbstract
Neisseria gonorrhoeae is a cause of gonorrhea which is the most common sexually transmitted diseases (STDs). Bacterial culture is a gold standard method for this bacterial detection, but it is time-consuming and skilled technicians are needed. Therefore, the development of a rapid, easy and sensitive detection technique is still required. In the present study, we developed a specific gold nanoparticle probe for the detection of N. gonorrhoeae genomic DNA compared to a conventional PCR method. The average size of the synthesized gold nanoparticles (GNPs) was approximately 13 nanometers. GNP probes could bind specifically with complementary sequences of DNA. Due to their salt-tolerance, GNPs solution remained in red color, which could be detected by naked eyes, after adding 100 mM MgSO4 solution. This method is easy and rapid with the detection limit of 20 ng/?L genomic DNA of N. gonorrhoeae. Twelve strains of bacteria were tested by this detection method and the results showed high specificity to N. gonorrhoeae. In conclusion, this developed method has the potential to apply for medical diagnosis.
Keywords
Neisseria gonorrhoeae, Medical diagnosis, Gold nanoparticles, DNA detection, DNA probe
Abbreviations
DNA: Deoxyribonucleic Acid, GNPs: Gold Nanoparticles, HPLC: High Performance Liquid Chromatographic technique, NAAT: Nucleic Acid Amplification Test, PBS: Phosphate Buffered Saline, PCR: Polymerase Chain Reaction, SDS: Sodium Dodecyl Sulfate, STDs: Sexually Transmitted Diseases, TEM: Transmission Electron Microscope.
Introduction
Gonorrhea is a sexually transmitted disease caused by Neisseria gonorrhoeae [1]. The rate of gonococcal infection is still one of the most common worldwide, even using the condom [2], because of untreatable multidrug-resistance [1]. Those who have asymptomatic infections with N. gonorrhoeae, with no treatment, may get many complications in future such as prostatitis, orchitis, ectopic pregnancy, abortion and conjunctivitis in foetus.
Methods for N. gonorrhoeae detection include bacterial culture, gram stain, conventional Polymerase Chain Reaction (PCR), hybridization, nucleic acid amplification test (NAAT) and serological test. Bacterial culture is a gold standard method for N. gonorrhoeae detection with great specificity, but also low sensitivity and time-consuming [3,4]. Gram-stained method has high sensitivity and specificity for the diagnosis of symptomatic men with urethral discharge but it is not appropriate for diagnosis of cervical, rectal and pharyngea gonorrhea, or for asymptomatic patients because of its low sensitivity [5]. PCR sensitivity and specificity depend on primer design [6]. NAAT is more sensitive than culture technique, especially for pharyngeal and rectal specimens, but inhibitory substances in patient specimens may cause the falsenegative results [7]. Hybridization method is a rapid and specific diagnosis of pathogens that are fastidious or cannot be cultured [6].
Taken together, we aimed to develop an accurate and rapid detection of N. gonorrhoeae by modification of hybridization technique to obtain higher sensitivity, cheap and rapid compared to culture technique. Specific DNA probe for N. gonorrhoeae conjugated with GNPs was designed for the specific sequence in the porB gene which codes for the major outer membrane protein (PorB) of N. gonorrhoeae. PorB can act as an anion-selective porin, interact with host cells, induce or inhibit apoptotic signaling in eukaryotic cells and has a relationship with human host pathogenesis [8,9]. The sensitivity and specificity of this method were also studied.
Materials and Methods
Bacterial strains and reagents
N. gonorrhoeae were obtained from the Clinical Microbiology Laboratory of Srinagarind Hospital, Khon Kaen University, Thailand. Reference strains of Klebsiella pneumonia, Corynebacterium spp., Pseudomonas aeruginosa, Staphylococcus epidermidis, Proteus mirabilis, Escherichia coli, Enterococcus spp., Staphylococcus aureus, Group B streptococcus, Listeria monocytogenes were obtained from Department of Microbiology, Faculty of Associated Medical Sciences, Khon Kaen University. HAuCl4.4H2O and sodium citrate dihydrate were purchased from Sigma-Aldrich Co., LLC (Missouri). The other chemicals were all of analytical grade.
Probe and primers design
The sequences of a DNA probe and two primers were designed from the porB gene. All oligonucleotides were generated as sequences using clustalW under the high homology among the N. gonorrhoeae. The NCBI BLAST (Basic Local Alignment Search Tool) search of the base target in porB gene showed highly specific for N. gonorrhoeae and no exact match against non-gonorrhoeae family members. Probes were labeled with thiol group (-SH) at 5’ end to enable conjugation with the GNPs surface. The probe and primer set were synthesized and then purified by Pacific Science Co., Ltd., Thailand.
N. gonorrhoeae detection by PCR method
The reaction mixtures of PCR were carried out in a final volume of 25 μL containing 50 mM KCl, 10 mM Tris-HCl pH 8.3, a 0.2 mM concentration of each dNTP, 2 mM MgCl2, 3 unit of thermostable DNA polymerase and 0.3 μM of each primer. The primary PCR amplification was performed using primers which listed in Table 1. The primary PCR program was preceded by a 4-min denaturation step at 94°C, followed by 40 cycles of 94°C for 1 min, 40°C for 1 min, and 72°C for 2 min, and the last cycle was followed by an additional 10-min elongation step at 72°C. PCR amplicons were analyzed by electrophoresis at 100 V in 1.5% agarose gels staining with ethidium bromide and visualized under UV light.
Preparation of gold nanoparticles
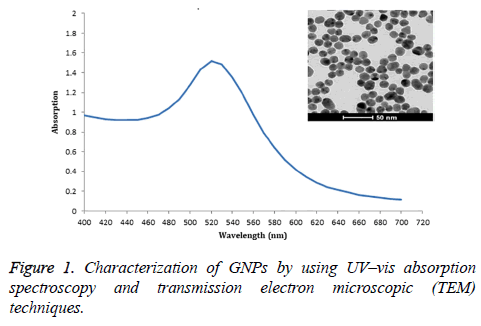
Gold nanoparticles (GNPs) were synthesized by the slight modification of the citrate reduction [10]. Briefly, 100 mL of 1 mM H-AuCl44H2O solution was mixed with 10 mL of a 38.8 mM trisodium citrate solution and refluxed for 15 min and then cooled down to room temperature. The morphology and size of the GNPs were characterized by using a field emission highresolution transmission electron microscope (TEM) (TECNAI G2 20 Model, Hillsboro, OR) operating at 200 kV. GNPs sample was prepared for TEM analysis by dropping them onto a carbon film copper grid and drying at room temperature. The result of TEM was analyzed with Image J (NIH, Bethesda, Maryland) for measuring the average size of GNPs. The absorption spectra (400-700 nm) were also measured using a SpectraMax M5 fluorescence microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA).
Preparation of oligo probe-conjugated GNPs
The conditions for labeling the GNPs with the N. gonorrhoeae oligonucleotide probes were modified from a previous study [10]. The reaction mixture of GNP probes was carried out at a final concentration of 0.50 and 1 μM oligonucleotide probes containing 1 mL of GNPs solution (GNP-0.5 μM probe and GNP-1 μM probe, respectively) and incubated on an orbital shaker overnight at 50°C. The phosphate buffer, SDS, and NaCl solution were added to obtain a final concentration of 10 mM phosphate buffer (pH 7.4), 0.01% (W/V) SDS, and 0.1 M NaCl. The mixture was incubated on an orbital shaker for 48 hours at 50°C, followed by centrifugation at 15,000 rpm 4°C for 30 min to remove unconjugated oligonucleotides. The GNP probes were washed with washing buffer (100 mM PBS containing 100 mM NaCl and 0.01% SDS) twice, then suspended in 200 μL of washing buffer and stored in dark at 4°C.
Estimation of the percentage of GNP probes
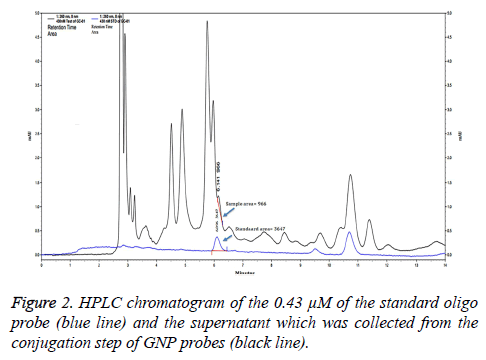
The concentration of unbound oligonucleotide probes after conjugation with GNPs was estimated by high-performance liquid chromatographic technique (HPLC) using an indirect assay [11]. After the reaction mixture of 1 mL of GNPs, probe (a final concentration of 0.50 μM oligonucleotide probe containing 1 mL of GNPs solution), phosphate buffer, SDS, and NaCl solution were performed; a final concentration of probe was 0.43 μM. The supernatant from the GNP probe conjugation step was injected into a Luna C18 (15 cm × 3.0 mm) HPLC column (Phenomenex, California). Gradient elution was conducted at a flow rate of 1.5 mL/min using 0.1% trifluoroacetic acid and 60% acetonitrile in 0.1% trifluoroacetic acid at ratios of 100:0, 80:20, and 0:100 at 0-5, 5-10, 15-25, and 30 min, respectively. The concentration of 0.43 μM of the oligo probe was used as a standard. Peak area corresponded to unbound probe in the supernatant was calculated with an oligonucleotide probe standard curve.
Genomic DNA extraction
The bacteria were cultured in chocolate agar at 37°C for 18-24 hours. Colonies were transferred to new tubes in 200 μL of deionized water and then boiled in boiling water for 10 min, then centrifuged at 13,000 rpm for 10 min and the supernatant was collected. The concentration of genomic DNA was quantified by measuring the optical density at 260 and 280 nm by using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The supernatant was kept at -20°C until use.
GNP probes hybridization assay
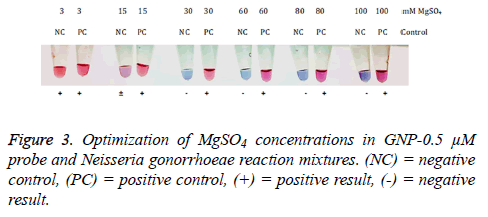
Optimization of salt concentration: The 3 μL of 50 mM of N. gonorrhoeae genomic DNA were added to 3 μL GNP probes. The mixtures were incubated at 95°C for 5 min, followed by 63°C for 5 min. After that MgSO4 at various concentration (3, 15, 30, 60, 80, 100 mM) was added. The positive result was red or pink and the negative result was blue solution which could be observed by naked eye. Deionized water was used as a negative control.
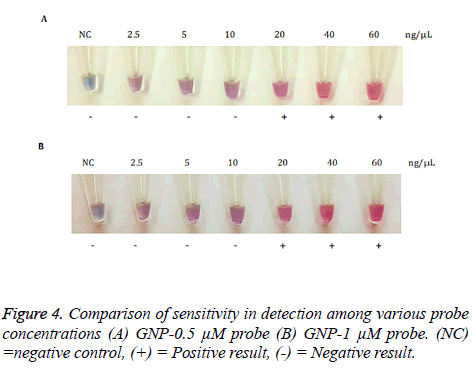
The sensitivity of GNP probes: N. gonorrhoeae genomic DNA was serially diluted to be 2.5, 5, 10, 20, 40, 60 ng/μL. Each genomic DNA concentration was hybridized with GNP probes. After the hybridization step, MgSO4 solution was added.
The specificity of GNP probes: Genomic DNA of N. gonorrhoeae and other 11 bacterial strains were diluted to final concentration of 50 ng/μL. All bacterial strains were hybridized with GNP probes. After the hybridization step, MgSO4 solution was added.
Results
Characterization of GNPs
These GNPs, prepared by sodium citrate reduction method, were spherical shape and size distribution about 13 nm diameter by TEM study. The maximum absorption of GNPs was 520 nm (Figure 1).
Estimation of the percentage of GNP probes
To evaluate the percentage of the GNP probes, the supernatant from the conjugation step of GNP probes was analyzed by HPLC and compared with a standard DNA probe (Figure 2). Peak area of unbound oligo probes and 0.43 μM of the standard oligo probes were 966 and 3647. The percentage of unbound oligo probes was 26.49%. Thus, the percentage of GNP probes was approximate 73.51%.
Optimization of salt concentration
After hybridization step, each of MgSO4 concentrations was added into mixtures containing GNP-0.5 μM probe and N. gonorrhoeae genomic DNA. The results of the reactions were compared between positive and negative control at different salt concentrations. The results showed that 30, 60, 80, and 100 mM MgSO4 can distinguish N. gonorrhoeae from negative control (Figure 3). The concentration of MgSO4 at 100 mM was selected as an optimum salt concentration.
The sensitivity of GNP probe
GNP probes were hybridized with 2.5, 5, 10, 20, 40, 60 ng/μL of N. gonorrhoeae genomic DNA by using sterile deionized water as a negative control (Figure 4). The solution containing 20, 40 and 60 ng/μL of N. gonorrhoeae genomic DNA remained red solution after MgSO4 was added, whereas the solution containing genomic DNA concentration less than 20 ng/μL and negative control solution turned into purple. The sensitivity in between GNP-0.5 μM probe and GNP-1 μM probe was not different. Therefore, GNP-0.5 μM probe was selected to evaluate the specificity of this method. The detection limit was 20 ng/μL genomic DNA.
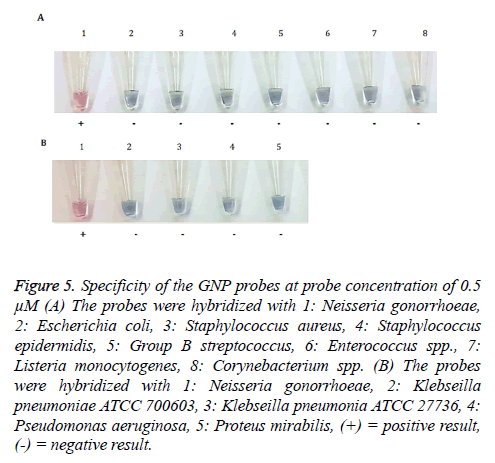
The specificity of GNP probes
To assess the specificity of GNP-0.5 μM probe, the probes were hybridized with 12 bacterial strains including N. gonorrhoeae. Only reaction containing N. gonorrhoeae genomic DNA turned into red solution, whereas other reactions with 11 bacterial strains turned into purple solution after addition of 100 mM MgSO4 (Figure 5).
Figure 5: Specificity of the GNP probes at probe concentration of 0.5 μM (A) The probes were hybridized with 1: Neisseria gonorrhoeae, 2: Escherichia coli, 3: Staphylococcus aureus, 4: Staphylococcus epidermidis, 5: Group B streptococcus, 6: Enterococcus spp., 7: Listeria monocytogenes, 8: Corynebacterium spp. (B) The probes were hybridized with 1: Neisseria gonorrhoeae, 2: Klebseilla pneumoniae ATCC 700603, 3: Klebseilla pneumonia ATCC 27736, 4: Pseudomonas aeruginosa, 5: Proteus mirabilis, (+) = positive result, (-) = negative result.
N. gonorrhoeae detection by PCR
In this study, the PCR technique was used as a reference method. We used specific primers to porB gene of N. gonorrhoeae and probe sequence was between the DNA sequence of PCR product. PCR products contained a band of 284 bp in length. The N. gonorrhoeae and other bacteria were examined by PCR assay to confirm the presence of the porB gene, which was the target gene of the GNP-probe assay. N. gonorrhoeae exhibited a band in gel electrophoresis at about 300 bp, whereas the others showed negative results (Figure 6). The result of PCR method was similar to the result of specificity by using GNPs-probe.
Figure 6: Agarose gel electrophoresis of PCR products. M: marker band, 1: Neisseria gonorrhoeae, 2: Escherichia coli, 3: Staphylococcus aureus, 4: Proteus mirabilis, 5: Listeria monocytogenes, 6: Group B streptococcus, 7: Staphylococcus epidermidis, 8: Corynebacterium spp., 9: Enterococcus spp., 10: Klebseilla pneumoniae ATCC 700603, 11: Klebseilla pneumonia ATCC 27736, 12: Pseudomonas aeruginosa.
Discussion
The incidence of gonorrhea are about 106 million adults each year worldwide, and infection rates in the developing countries are about 10-20 times more than in the developed countries [12]. Although bacterial culture is a gold standard method to identify N. gonorrhoeae infection but it has low sensitivity and time-consuming [12]. The other methods also have both advantages and disadvantages [5-7] Therefore, we develop a novel detection method for diagnosis of N. gonorrhoeae. A previous study reported that colorimetric assay using GNPs conjugated with thiol-modified oligonucleotide probes are rapid and the product result from this reaction can be observed with the naked eye without complicated and expensive instrumentation [13]. The colorimetric using GNPs-probe assay is faster than culture method and requires only a thermal block. Forming the linkage of thiol-ssDNA and GNPs for gold nanoprobe assay is easy, inexpensive in common molecular assays, and this solution can be kept at room temperature for 1 month [14,15]. In the present study, we used ~13 nm diameter GNPs for the colorimetric detection of DNA because the result of hybridization can be easily detected visually [11,16]. The hybridization of GNPs-probe with the target DNA formed double-stranded DNA helix that made GNPs more salt-tolerant than single-stranded DNA (No target DNA). Therefore, the hybridization of GNPs-probe with the target DNA showed red solution. The concentration of MgSO4 was optimized after hybridization. A 100 mM MgSO4 was used in this study due to the specificity test of GNPs-probe colorimetric assay, genomic DNA of N. gonorrhoeae and some other bacteria could not be discriminated by naked eye at less than 100 mM MgSO4 (data not shown). Therefore, some concentration of MgSO4 can distinguish between negative control (deionized water) and N. gonorrhoeae but not between bacterial strains. The detection limit of our study was 20 ng/μL unamplified genomic DNA which is better than a previous study that the limit of detection was ~54 ng/μL [17]. The different detection limit may arise from the different preparation step of GNPs-probe. The developed GNPs-probe assay had specificity to N. gonorrhoeae DNA. The result of PCR method was related to the result of GNP probe colorimetric assay in term of specificity and can confirm that positive control is true.
Conclusion
The present study reports a novel method for Neisseria gonorrhoeae genomic DNA detection based on GNP probe colorimetric assay. This developed method can distinguish positive and negative control by naked eye. The detection limit of this method was 20 ng/μL genomic DNA. It is a rapid and easy method with high sensitivity and specificity. This method also has a potential method to apply for medical diagnosis.
Acknowledgement
This work was supported by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission through the Center of Excellence in Specific Health Problems in the Greater Mekong Sub-region cluster (SHeP-GMS), Khon Kaen University, Thailand. (.)
Conflict of Interest
The authors declare that there are no conflicts of interest.
References
- Warner L, Stone KM, Macaluso M, Buehler JW, Austin HD. Condom use and risk of gonorrhea and Chlamydia: a systematic review of design and measurement factors assessed in epidemiologic studies. Sex Transm Dis 2006, 33:36-51.
- Craig AP, Gray RT, Edwards JL, Apicella MA, Jennings MP, Wilson DP, Seib KL. The potential impact of vaccination on the prevalence of gonorrhea. Vaccine 2015, 33:4520-4525.
- Van Dyck E, Ieven M, Pattyn S, Van Damme L, Laga M. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by enzyme immunoassay, culture, and three nucleic acid amplification tests. J Clin Microbiol 2001, 39:1751-1756.
- Lewis DA. Will targeting oropharyngeal gonorrhoea delay the further emergence of drug-resistant Neisseria gonorrhoeae strains? Sex Transm Infect 2015, 91:234-237.
- Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014, 27:587-613.
- Ng LK, Martin IE. The laboratory diagnosis of Neisseria gonorrhoeae. Can J Infect Dis Med Microbiol 2005, 16:15-25.
- Whiley DM, Tapsall JW, Sloots TP. Nucleic acid amplification testing for Neisseria gonorrhoeae: an ongoing challenge. J Mol Diagn 2006, 8:3-15.
- Chen A, Seifert HS. Structure-function studies of the Neisseria gonorrhoeae major outer membrane porin. Infect Immun 2013, 81:4383-4391.
- Chen A, Seifert HS. Saturating mutagenesis of an essential gene: a majority of the Neisseria gonorrhoeae major outer membrane porin (PorB) is mutable. Bacteriol 2014, 196:540-547.
- Phromjai J, Mathuros T, Phokharatkul D, Prombun P, Suebsing R, Tuantranont A, Kiatpathomchai W. RT-LAMP detection of shrimp Taura syndrome virus (TSV) by combination with a nanogold-oligo probe. Aquac Res 2015, 46:1902-1913.
- Kumvongpin R, Jearanaikool P, Wilailuckana C, Sae-Ung N, Prasongdee P, Daduang S, Wongsena M, Boonsiri P, Kiatpathomchai W, Swangvaree SS, Sandee A, Daduang J. High sensitivity, loop-mediated isothermal amplification combined with colorimetric gold-nanoparticle probes for visual detection of high risk human papillomavirus genotypes 16 and 18. J Virol Methods 2016, 234:90-95.
- Liu ML, Xia Y, Wu XZ, Huang JQ, Guo XG. Loop-mediated isothermal amplification of Neisseria gonorrhoeae porA pseudogene: a rapid and reliable method to detect gonorrhea. AMB Express 2017, 7:48.
- Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev 2005, 105:1547-1562.
- Costa P, Amaro A, Botelho A, Inacio J, Baptista PV. Gold nanoprobe assay for the identification of mycobacteria of the Mycobacterium tuberculosis complex. Clin Microbiol Infect 2010, 16:1464-1469.
- Hill HD, Mirkin CA. The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat Protoc 2006, 1:324-336.
- Dykman LA, Khlebtsov NG . Gold nanoparticles in biology and medicine: recent advances and prospects. Acta Naturae 2011, 3:34-55.
- Bakthavathsalam P, Rajendran VK, Mohammed JAB. A direct detection of Escherichia coli genomic DNA using gold nanoprobes. J Nanobiotechnology 2012, 10:8.