- Biomedical Research (2014) Volume 25, Issue 1
Rapid Dissemination of Multidrug Resistant Providencia Stuartii- A University Hospital Based Study.
Bahadır Feyzioğlu1, Dilek Güldemir2, Alper Karagöz2, İbrahim Erayman3, Mehmet Emin Demircili1 and Mahmut Baykan11Department of Medical Microbiology, Necmettin Erbakan University Meram Faculty of Medicine, Konya, Turkey
2Molecular Microbiology Research and Application Laboratory, Department of Microbiology Reference Laboratories, Public Health Agency of Turkey, Ankara, Turkey
3Department of Infectious Diseases, Necmettin Erbakan University Meram Faculty of Medicine, Konya, Turkey
- *Corresponding Author:
- Bahadır Feyzioğlu
Department of Medical Microbiology
Necmettin Erbakan University
Meram Faculty of Medicine
Konya, Turkey
Accepted date: November 18 2013
Abstract
Providencia stuartii infections are not common. There are a few reports of declared outbreaks or spreads. In our study, nosocomial dissemination by extended-spectrum β-lactamase (ESBL) producing Providencia stuartii in a variety of clinics was investigated in a short period. Nine multidrug-resistant Providencia stuartii clinical isolates were collected between October and November in 2011. Nine isolates were obtained from six patients who had been hospitalized in a variety of clinics at a university hospital in Turkey. The clonal relationship among nine isolates was determined by pulsed-field gel electrophoresis. All isolates were ESBL positive and resistant to quinolones, and we determined amikacin resistance to three strains. There were two antibiotypes, and a second PFGE profile was not observed in different types. The outbreak was due to the dissemination of one epidemic multiresistant clone of Providencia stuartii. We considered that it had been a common dissemination from a single source and the first isolate which was collected from the first patient was epidemic clone. The respiratory tract colonization of the first patient was considered to be the possible route of transmission. The resistance to amikacin occurred during treatment with aminoglycoside. It was noteworthy.
Keywords
ESBL, multidrug resistance, nosocomial outbreak, Providencia stuartii, pulsed field gel electrophoresis
Introduction
The genus Providencia is responsible for a wide range of human infections, but they are rare and usually nosocomial. The first species of the genus now known as Providencia was isolated by Rettger in 1904. There are 5 species of the genus Providencia. In descending order of prevalence, they include Providencia stuartii, Providencia rettgeri, Providencia alcalifaciens, Providencia rustigianii and Providencia heimbachae [1].
Providencia stuartii is a gram stain negative, facultative anaerobe and motile bacteria. It is found in multiple animal reservoirs such as flies, birds, cats, dogs, cattle, sheep and is also found commonly in soil, water, and sewage. It is an opportunistic pathogen seen in patients with severe burns or long-term indwelling urinary catheters. Providencia stuartii outbreaks are usually of urinary origin. This species has been isolated from urine, stool, and blood, as well as from sputum, skin, and wound cultures. On the other hand, Providencia infections represent an emerging problem because of the increasing prevalence of antibiotic resistance. Providencia stuartii is an antibiotic multi-resistant bacterium of the genus. Especially ESBLpositive strains are a problem in hospitalized patients [2,3]
Providencia infections are not common. There are a few reports declared outbreak or spread. In our study, nosocomial dissemination by extended-spectrum β-lactamase (ESBL)-producing Providencia stuartii in a variety of clinics was investigated in a short period (one month). This is the first study reported in this sense in our country. Moreover, there are a limited number of reports in this regard globally.
Materials and Methods
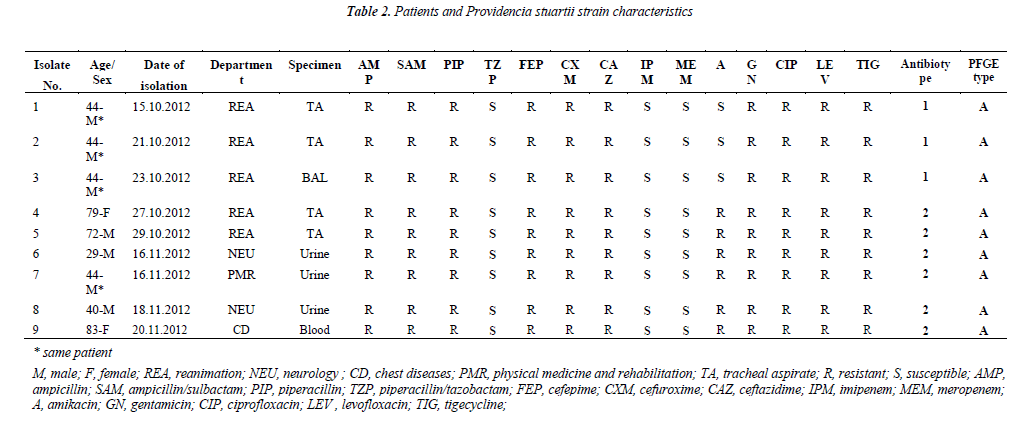
Nine multidrug-resistant Providencia stuartii clinical isolates were collected between October and November in 2011. Nine isolates were obtained from six patients who had been hospitalized in a variety of clinics (reanimation, physical medicine and rehabilitation, neurology, and chest diseases) at the university hospital of Turkey. These isolates were obtained from culture of urine, tracheal aspiration, broncho alveolar lavage (BAL) and blood specimens.
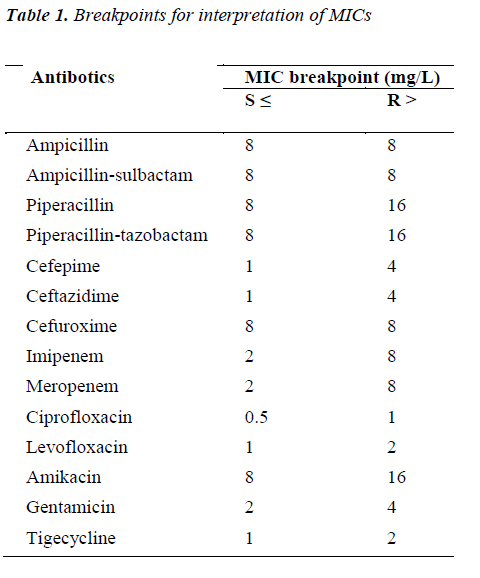
Isolates were identified by using either automated Vitek 2 system (BioMérieux) or conventional methods. Antimicrobial susceptibility testing was performed using the disk diffusion method on Mueller-Hinton agar plates. Minimal inhibitory concentrations (MIC) were determined by the agar dilution technique and interpretations were made according to EUCAST (The European Committee on Antimicrobial Susceptibility Testing) criteria (Table 1) [4]. ESBL detection was performed by the double-disk synergy test as previously described [5].
The specimens were immediately collected for culture from all patients (urine, blood, tracheal aspiration or broncho alveolar lavage, rectal swabs, axilla swabs, and nose swabs) and the environment (such as, aspirator probe, aspirator reservoir, head of the bed, the nurse desk, on the ground, tap water) in the reanimation department, after first Providencia strain was isolated. Then, Providencia stuartii was isolated from tracheal aspiration of the first patient but was not isolated from other specimens. Subsequently, we defined Providencia stuartii from a variety of routine samples collected from the same patient and other patients in different departments (physical medicine and rehabilitation, neurology, and chest diseases clinic). In order to find the contamination source in these departments, we did an investigation as defined above, but the bacteria were not determined from the collected specimens.
A total of nine isolates that were obtained from six patients who had been hospitalized in a variety of clinics were identified as Providencia stuartii in a one-month period.
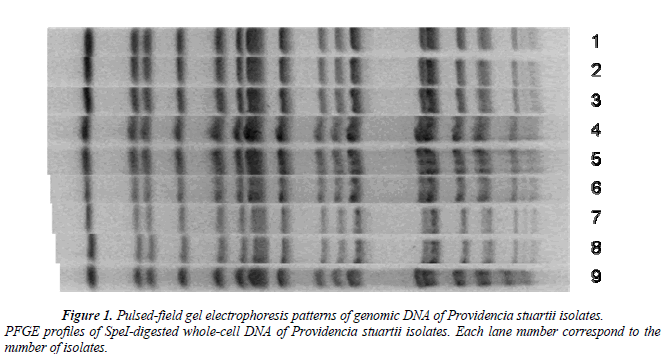
The clonal relationship among nine isolates was determined by pulsed-field gel electrophoresis (PFGE) as previously described. PFGE was performed using the SpeI restriction enzyme (10 U/μl) (Promega Corporation, WI, USA) and buffer. A contour–clumped homogeneous electric field (CHEF) method was used for PFGE on a CHEF DRII system (Bio-Rad). The dendogram was obtained by Bionumerics program.
Results
A total of nine isolates that were obtained from six patients who had been hospitalized in a variety of clinics were identified as Providencia stuartii in a one-month period. Four (two tracheal aspiration, urine and BAL) of them were isolated from the patient from whom the first isolation was determined (Table 2).
All isolates were highly resistant to beta-lactams, aminoglycosides, and fluoroquinolones, except meropenem, imipenem, and piperacillin/tazobactam. All isolates of Providencia stuartii were ESBL positive and resistant to quinolones. We determined amikacin resistance except in three strains which were isolated from the same patient. So, two different antibiotypes were determined (Table 2).
All the Providencia stuartii isolates yielded one PFGE profile. Although there were two antibiotypes, a second PFGE profile was not observed in different types (Figure 1).
Moreover, similar isolates have not been recovered before or after the two-month survey period.
Discussion
Providencia stuartii is aetiological agents of nosocomial infections. It increases especially in the patients who recive medical care. It often leads to urinary tract infections, and causes, though less frequently, respiratory and skin infections. There are predisposing factors such as catheterization and intubation causing infections with this bacterium. Recently reported studies have expressed that the increase in infections of Providencia stuartii is associated with the increased use of some antibiotics such as colistin [6]. These species have been isolated from urine, stool, and blood, as well as from sputum, skin, and wound cultures. We have isolated nine Providencia stuartii strains from blood, sputum, and tracheal aspirate specimens. Providencia stuartii is the least susceptible Providencia sp. and is naturally resistant to tetracyclines, some penicillins, older cephalosporins, sulphamethoxazole and fosfomycin [7]. However, the acquired resistance is an increasing problem. Providencia stuartii can be multiresistant to antibiotics especially to beta-lactams. Nevertheless, considering reports up to date, multi-drug resistance was not a common situation. There are limited numbers of notification related to outbreaks or dissemination caused by multidrug resistant Providencia species [3,8,9,10]. In our study, nosocomial dissemination caused by Providencia stuartii occurred in a period of one month. Providencia stuartii had an extended-spectrum β- lactamase (ESBL) and was resistant to quinolones and amikacin. This research is the first study on this topic in our country and there are limited studies in the world.
In this study, antibiotic susceptibility profiles of the strains were determined and determination of genotype profiles was aimed at examining the clonal relationship.
We identified two different antibiotypes. Presence of ESBL and resistance to quinolones were detected in all the isolates, whereas amikacin resistance was determined in all, except three strains which were isolated from the same patient. Four isolates were obtained from this patient within one month. The first three of these four isolates were susceptible to amikacin, whereas the last isolate was resistant to amikacin and it was considered to be the second antibiotype. First antibiotype complies with (2'-N-acetyltransferase), AAC (2') and the second antibiotype complies with (6'-N-acetyltransferase type) AAC (6 ') aminoglycoside resistance profile. The characterization of AAC (2') is resistant to all aminoglycosides except of kanamycin and amikacin. AAC (6') is an enzyme with activity responsible for amikacin resistance in gramnegative bacteria, especially in the family Enterobacteriaceae [11,12]. Some studies showed a similarity between the AAC (2') and AAC(6') promoters, and emphasized the potential of mutation in Enterobacteriaceae [13]. Another study showed that the AAC (2') in some strains of Providencia stuartii can be induced to high levels of activity by exposure to aminoglycosides. In this study, we determined that resistant to amikacin may be associated with this similarity or mutations and may be the subject of another study. Also, the second antibiotype profile was determined from five isolates obtained from other patients. We determined that amikacin treatment had been applied to the patient at the date range (08th-29th of October) when the first three strains were obtained.
The infections caused by ESBL-producing strains of Enterobacteriaceae are increasing day by day, but fewer numbers of cases of ESBL producing Providencia stuartii infections have been reported to date [14]. Otherwise, prevalence and phenotypic characteristics of ESBL producing Providencia stuartii infections exhibit variation among geographical areas. The reports in publications particularly in countries such as Tunisia, Italy and the Balkan countries have been noteworthy in recent years [8,9,14,17,18]. Multi-strain work has not been reported except for a few case reports in our country [20,21].
In our study, we used PFGE to assess a dissemination of ESBL producing multidrug resistant Providencia stuartii which occurred in a variety of clinics at a university hospital. The results of typing analysis suggest that the outbreak was mainly due to the dissemination of one epidemic multiresistant clone of Providencia stuartii. We considered that it had been a common dissemination from a single source and the first isolate which was collected from the first patient was epidemic clone. The respiratory tract colonization of the first patient was considered to be the possible route of transmission. A well-known fact is that many bacteria transmitted during healthcare derive from human sources and inanimate environmental sources [22]. In this regard, vector can be healthcare involving touching the contaminated equipment before managing the patient. By considering these situations, we collected samples from all associated patients and inanimate environments after the first isolation. Almost no bacteria were obtained from these specimens, except routine patient specimens. After the spread of the first isolated bacteria to the environment, the dissemination to other patients may have occurred by healthcare involving touching the contaminated equipment before managing the patient. It is remarkable that one epidemic clone of multi-resistant Providencia stuartii in different clinics was detected, although patients and their services were not related to each other. We considered that this quick dissemination in such a short period of time may have been caused by factors such as the hospital surfaces and contaminated water, etc. However, alternative ideas for route of transmission are open to debate.
In our study, nosocomial dissemination caused by Providencia stuartii occurred in a period of one month was investigated. There was an ESBL producion and resistance to quinolones and amikacin. This is the first report related to Providencia stuartii dissemination in our country. Two antibiotypes and a single genotype were determined for the nine strains. The occurrence of resistance to amikacin in common clonal strains during treatment with aminoglycoside was considered important information. The outbreak was monoclonal and transmitssion of epidemic Providencia stuartii isolates was through a common source. The respiratory tract of the first patient was considered to be the source. We must be ready for new outbreaks which may lead to similar multidrug resistant strains.
References
- Manos J, Belas R. The Genera Proteus, Providencia, and Morganella. Prokaryotes 2006; 6: 245-269.
- Whiteley GR, Penner JL, Stewart IO, Stokan PC, Hinton NA. Nosocomial urinary tract infections caused by two O-serotypes of Providencia stuartii in one hospital. J Clin Microbiol 1977; 6: 551-554.
- Kocka FE, Srinivasan S, Mowjood M, Kantor HS. Nosocomial multiply resistant Providencia stuartii: a long-term outbreak with multiple biotypes and serotypes at one hospital. J Clin Microbiol 1980; 11: 167-169.
- The European Committee on Antimicrobial Susceptibility Testing. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance [cited 2012 December]. Available from: http://www.eucast.org
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 3.1, 2013. [cited 2012 December]. Available from: www. eucast. org
- Hayakawa K, Marchaim D, Divine GW, Pogue JM, Kumar S, Lephart P, Risko K, Sobel JD, Kaye KS. Growing prevalence of Providencia stuartii associated with the increased usage of colistin at a tertiary health care center. Int J Infect Dis 2012; 16: 646-8
- Stock I, Wiedemann B. Natural antibiotic susceptibility of Providencia stuartii, P. rettgeri, P. alcalifaciens and P. rustigianii strains. J Med Microbiol 1998;47: 629-642.
- Saida NB, Thabet L, Messadi A, Bouselmi K, Turki A, Boukadida J. Clonality of Providencia stuartii isolates involved in outbreak that occurred in a burn unit. Burns 2008; 34: 829-834.
- Lahlaoui H, Poirel L, Moussa MB, Ferjani M, Omrane B, Nordmann P. Nosocomial dissemination of extended-spectrum β-lactamase VEB-1a-producing Providencia stuartii isolates in a Tunisian hospital. Eur J Clin Microbiol Infect Dis 2011; 30: 1267-1270.
- Arpin C, Thabet L, Yassine H, Messadi AA, Boukadida J, Dubois V, Coulange-Mayonnove L, Andre C, Quentin C. Evolution of an incompatibility group IncA/C plasmid harboring blaCMY-16 and qnrA6 genes and its transfer through three clones of Providencia stuartii during a two-year outbreak in a Tunisian burn unit. Antimicrob Agents Chemother 2012; 56: 1342-1349
- Franklin K, Clarke AJ. Overexpression and characterization of the chromosomal aminoglycoside 2'-Nacetyltransferase of Providencia stuartii. Antimicrob Agents Chemother 2001; 45: 2238-2244
- Lambert T, Gerbaud G, Courvalin P. Characterization of transposon Tn1528,which confers amikacin resistance by synthesis of aminoglycoside 3'-Ophosphotransferase type VI. Antimicrob Agents Chemother 1994; 38: 702-706
- Rather PN, Orosz E, Shaw KJ, Hare R, Miller G. Characterization and transcriptional regulation of the 2'-N-acetyltransferase gene from Providencia stuartii. J Bacteriol 1993; 175: 6492-6498.
- Barl P, Bedenić B, Sardelić S, Uzunović S, Vraneš J, Plečko V. Spread of CTX-M-15 positive Providencia spp. causing urinary tract infections at the University Hospital Split in Croatia. Med Glas (Zenica) 2012; 9: 317-324
- Bonfiglio G, Perilli M, Stefani S, Amicosante G, Nicoletti G. Prevalence of extended spectrum betalactamases among Enterobacteriaceae: an Italian survey. Int J Antimicrob Agents 2002; 19: 213-217.
- Tumbarello M, Citton R, Spanu T, Sanguinetti M, Romano L, Fadda G, Cauda R. ESBL-producing multidrug-resistant Providencia stuartii infections in a university hospital. J Antimicrob Chemother 2004; 53: 277-282.
- Mnif B, Ktari S, Chaari A, Medhioub F, Rhimi F, Bouaziz M, Hammami A. Nosocomial dissemination of Providencia stuartii isolates carrying bla OXA-48,bla PER-1, bla CMY-4 and qnrA6 in a Tunisian hospital. J Antimicrob Chemother 2013; 68: 329-332.
- Arnoldo L, Migliavacca R, Regattin L, Raglio A, Pagani L, Nucleo E, Spalla M, Vailati F, Agodi A, Mosca A, Zotti C, Tardivo S, Bianco I, Rulli A, Gualdi P, Panetta P, Pasini C, Pedroni M, Brusaferro S. Prevalence of urinary colonization by extended spectrum-beta-lactamase Enterobacteriaceae among catheterised inpatients in Italian long term care facilities. BMC Infect Dis 2013; 13: 124
- Ayhan Tekiner, Zeliha Kocak Tufan, Mehmet Akif Bayar, Tuncer Tascioglua, Yavuz Selim Erkoca, Cigdem Hatipoglu. Nosocomial Providencia stuartii Meningitis: A Case Report. J Neurol Res 2011; 1: 37-39
- Sipahi OR, Bardak-Ozcem S, Ozgiray E, Aydemir S, Yurtseven T, Yamazhan T,Tasbakan M, Ulusoy S. Meningitis due to Providencia stuartii. J Clin Microbiol 2010; 48: 4667-4668
- Nazik H, Bektöre B, Öngen B, Özyurt M, Baylan O, Haznedaroğlu T. Co-expression of plasmid-mediated quinolone resistance-qnrA1 and blaVEB-1 gene in a Providencia stuartii strain. New Microbiol 2011; 34: 225-228
- Siegel JD, Rhinehart E, Jackson M, Chiarello L, and the Healthcare Infection Control Practices Advisory Committee, 2007 Guideline for Isolation Precautions: Preventing Transmission of Infectious Agents in Healthcare Settings. [cited 2013 June]. Available from: http://www.cdc.gov/ncidod/dhqp/pdf/isolation2007.pdf