Research Article - Journal of RNA and Genomics (2015) Research Report
Properties and kinetics of microRNA regulation through canonical seed sites
| Jerry S Chen1,3, Arra C Revilla3, Michael Guerrero3, Abygail M Gumbayan3 and Robert W Zeller1,2,3* 1Computational Science Research Center 2Center for Applied and Experimental Genomics 3Department of Biology, San Diego State University, 5500 Campanile Drive, San Diego, CA 92182, USA |
| Corresponding Author: Robert Zeller, E-mail: rzeller@mail.sdsu.edu, Tel: +619 594 6458, Fax: +619 594 5676 |
| Received: 26 November 2014; Revised: 22 January 2015; Accepted: 16 February 2015; Published: 16 February 2015 |
| © Copyright The Author(s). First Published by Library Publishing Media. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/ by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details. |
Abstract
MicroRNAs are a fundamental class of small RNAs involved in post-transcriptional gene regulation; however, the mechanism by which microRNAs regulate their gene targets in animals remains poorly understood. Practically, a mechanistic understanding of microRNA binding and regulation is crucial for the rational design of microRNA-based vectors for RNA interference. In this report, we focus on the largest known class of micro- RNA targets, the canonical seed targets, and explore the factors involved in modulating target downregulation in vivo at the protein level. Using an in vivo sensor assay in the ascidian Ciona intestinalis, we quantify miR-124-mediated downregulation of 38 canonical seed targets cloned from the Ciona genome as well as 10 control non-targets. Supporting previous findings, we observed that the seed type and number of seed sites are correlated with downregulation. However, up to a 50% variation in downregulation levels was observed for targets within the same seed class, indicating a role of non-seed factors in modulating downregulation. Although we did not observe a significant correlation of previously reported non-seed determinants with downregulation levels at saturation in our assay, our data suggest that two previously identified factors, secondary structure and 3’end complementarity, may play a role in the initial kinetics of microRNA-target binding. Importantly, using different concentrations of miR-124 we show that dose-dependent target downregulation profiles follow Michaelis-Menten kinetics. In summary, our findings emphasize the importance of non-seed factors as well as the importance of cellular concentrations of microRNAs relative to their targets when studying the mechanisms of endogenous microRNA regulation.
KEYWORDS |
| microRNA, miRNA targets, canonical target, seed site, Michaelis-Menten |
INTRODUCTION |
| MicroRNAs (miRNAs) are small RNAs that regulate gene expression and play critical roles in the development of most animals (Bartel, 2009; Pasquinelli, 2012). MiRNA regulation is also critical for proper homeostasis of adult tissues, as over half of all known human miRNAs have been implicated in some form of cancer or disease (Jiang et al, 2009). MiRNAs regulate gene expression by acting as guide RNAs that recruit RNA-induced silencing complexes (RISCs) to target transcripts, with Argonaute being the primary protein component that interacts with the target ( Pasquinelli, 2012). |
| This complex has sometimes been referred to collectively as the miRISC (Pasquinelli, 2012). After base pairing of miRNAs to target transcripts, the miRISC induces transcript degradation and/or translational inhibition, resulting in downregulation of gene expression (Bartel, 2009; Pasquinelli, 2012). |
| The precise mechanism of how miRNAs bind and regulate their target transcripts has been a subject of intense research over the past ten years. Although much is still uncertain, researchers agree that the 5’ seed region of the mature miRNA is important for target binding and robust downregulation (Brennecke et al, 2005; Bartel, 2009; Kim et al, 2009; Pasquinelli, 2012), of which there are three ‘canonical’ seed types that represent the major class of miRNA targets (Bartel, 2009). Many other factors such as 3’end complementarity, site location and secondary structure have also been suggested to be involved in miRNA target downregulation (Grimson et al, 2007; Kertesz et al, 2007), although only a limited number of studies support these findings and therefore warrant further experimental verification. Other species derived from the precursor miRNA such as the strand opposite the mature miRNA, called the miRNA*, and strands flanking the mature and miRNA*, called moRNAs, have also been implicated in some cases to have regulatory activity, also these cases also warrant further investigation (Shi et al, 2009; Yang et al, 2011). |
| In recent years, the endogenous microRNA pathway has been utilized as a vehicle for targeted silencing of gene expression through RNA interference (RNAi) (Borel et al, 2011; Chen and Zeller, 2014; Stegmeier et al, 2005). MiRNA-based RNA interference has several advantages over other methods such as introduction of long doubles-tranded RNA or RNA polymerase III-driven short hairpin RNAs, and holds tremendous promise as an effective gene knockdown strategy. However, one of the challenges with miRNA-based RNAi in animals is designing the miRNA vector to maximize target expression knockdown while minimizing off-target effects. While miRNA-based RNAi vectors are usually designed to target a specific site with full complementarity of the miRNA to the target transcript, the efficacy of seed and non-seed binding, as well as additional factors such as site location, secondary structure, and miRNA/target dosage, remains largely unknown. Therefore, a better understanding of how miRNAs target gene transcripts in vivo, which we explore in this report, will aid in the rational design of miRNA-based RNAi vectors. |
| Here, we use an in vivo miRNA target reporter assay developed in our laboratory to investigate determinants of miRNA targeting through canonical seed sites (Chen et al, 2011). The binding sites for these gene targets, for which the vast majority have been identified in transcript 3’untranslated regions (3’UTRs), contain a canonical base pairing of 6–8 nucleotides to the miRNA 5’ seed region (Bartel, 2009). We also investigate how other factors such as local sequence context, secondary structure and miRNA concentration affect targeting. We perform our experiments in the ascidian Ciona intestinalis, which we have previously shown contains the endogenous machinery necessary to process miRNA hairpin transcripts (Chen and Zeller, 2009; Chen et al, 2011; Tang et al, 2013). |
MATERIALS AND METHODS |
| Live animal sensor assay |
| Epidermally-driven (EpiB) miR-124 and reporter sensors were designed as described previously (Chen et al, 2011). Target constructs were PCR amplified from genomic DNA, restriction digested with Eco RI and Bgl II, and ligated into EpiB::RFP sensors. The endogenous genomic locus from which the miR-124 construct is derived contains two tandem copies of the miR-124 hairpin residing within the second intron of the host gene Ci-Pans. Since regions flanking the hairpin are known to be important for Drosha processing (Kim et al, 2009), we amplified the entire intron containing the miR-124 locus as well as the two flanking exons of the host gene. We verified previously that this construct produces mature miR-124 (Chen et al, 2011). Electroporations, image analysis and calculation of percent downregulation were performed as described previously (Zeller et al, 2006; Chen and Zeller, 2009; Chen et al, 2011). Unless otherwise stated, exactly 8g of each of the relevant DNA transgenes was electroporated into embryos. All of our electroporations were 200μl final volume in 4mm gap cuvettes. |
| Calculation of pooled standard deviation |
| Improving on our previous study (Chen et al, 2011), we developed a more accurate method for calculating the pooled standard deviation of percent downregulation. Given that z = 100%*(1-y/x) is the mean percent downregulation, where y is the mean normalized target RFP fluorescence and x is the mean normalized control RFP fluorescence, we can consider y/x as a new statistical parameter defined to be the ratio of the means of y and x. A biased measure of the variance of this ratio estimator, which has been used previously as a measurement of variance in fluorescence ratio imaging (van Kempen and Vliet, 2000), is given by: |
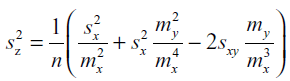 |
| where sx and sy are the standard deviations for the sets of control and target embryo fluorescence, respectively, mx and my are the respective means, and sxy is the covariance between control and target sets. Since the control and target sets are independent, sxy = 0. Although this estimator is biased, it is accurate to O(n-2), and, given our large sample size (average n=37 embryos per experiment), provides a reasonable estimate of the variance in downregulation. |
| Analysis of previously reported determinants of micro- RNA target downregulation |
| Ciona 3’UTR annotations are based on the most recent KH gene models (Satou et al, 2008). The location score is determined by the relative location of the target site within the 3’UTR, with higher scores for sites residing near the ends of the 3’UTR. The AU-context score is determined by the density of A:U base pairs immediately flanking the target site. The location score, AU-context score and 3’ non-seed complementarity score were calculated exactly as in Grimson et al (2007) and normalized by the maximum score. ΔGopen (dGopen) is a measure of the energy required to make a target site accessible for miRNA binding and is thus a direct measure of the ‘openness’ of the secondary structure surrounding the target site, while ΔΔG (ddG) is a measure of the change in total free energy upon miRNA binding (Kertesz et al, 2007). Both parameters were calculated using PITA (Kertesz et al, 2007) and Vienna RNA (Gruber et al, 2008). For each target site, we calculated the miRNA:target RNA duplex minimum free energy using RNAhybrid (Krüger and Rehmsmeier, 2006), allowing a maximum 6 nucleotide loop/bulge size, and allowing G:U wobble pairing. All other parameters were set as default values. Linear regression of each determinant against percent downregulation was carried out by performing an ANOVA test. P-values were calculated using t-tests of the slope parameter for each regression. |
| Model fitting |
| The ‘lm’ package was used in R to fit the titration data to a linear model, and the ‘drm’ package to fit the data to a specific non-linear model. For example, if the miR-124 dose is in column 1 of the data frame f and the target downregulation levels are in column 2, then the following commands were used: |
| Linear: mydata <- lm(f$V2~f$V1,data=f) |
| Logistic: mydata <- drm(f$V2~f$V1,data=f, fct=L.3(fixed=c(NA,NA,NA))) |
| Michaelis-Menten: mydata <- drm(f$V2~f$V1, data=f, fct=MM.2()) |
| Exponential: mydata <- drm(f$V2~f$V1, data=f, fct=EXD.2()) |
RESULTS AND DISCUSSION |
In vivo sensor assay for quantifying microRNA target downregulation |
| We developed an in vivo fluorescent reporter-based method for assaying microRNA target downregulation in Ciona embryos, which we used previously to verify predicted miR- 124 targets with conserved functions in neuronal development (Chen et al, 2011). The assay utilizes an electroporation technique developed by our laboratory that can introduce a plasmid DNA construct simultaneously into hundreds of live Ciona embryos in a single experiment (Zeller et al, 2006). In this report, we use this in vivo sensor assay to quantify relative downregulation of target gene expression at the protein level for dozens of gene targets (Figure 1A-B). We co-express precise levels of a vector containing the two contiguous copies of the miRNA miR-124 found in the endogenous genomic locus along with control and target fluorescent reporter constructs in Ciona epidermal tissues (see Materials and Methods). We focus on the largest known class of miRNA targets, the ‘canonical seed’ targets, which are estimated to represent at least two-thirds of all miRNA targets (Bartel, 2009). Canonical seed targets contain one of three possible ‘seed sites’ in their transcript 3’UTRs: (1) perfect complementarity to nucleotides 1–8 of the targeting miRNA (8mer), (2) complementarity to nucleotides 2–8 of the targeting miRNA (7mer-m8), and (3) complementarity to nucleotides 2–7 of the targeting miRNA with an ‘A’ opposite to position 1 of the miRNA (7mer-A1) (Bartel, 2009). Although rare instances of miRNA regulation in target coding regions have been verified (Pasquinelli, 2012), our focus here is on canonical 3’UTR targets, and therefore in our in vivo assay we cloned only the 3’UTR of predicted gene targets into reporter constructs (Figure 1A-B). In addition to quantifying 25 previously validated miR-124 targets cloned from the Ciona genome (Chen et al, 2011), for this report we assayed and quantified another 13 predicted canonical miR-124 targets in Ciona as well as an additional 9 control non-target genes, which do not contain canonical 3’UTR seed sites (Figure 1C and Table S1; previously validated targets indicated with an asterisk next to gene ID). For each transcript 3’UTR, we compared the relative reporter expression levels in the presence versus absence of ectopic miR- 124, comparing an average of n=74 pairs of Ciona embryos. All experiments were performed at least in duplicate with the exception of two non-target genes (Table S1). Finally, we pooled the calculated variance from all replicates for a given experiment to calculate a pooled standard deviation for each target. |
Gene target downregulation via the canonical 3’UTR seed site |
| Overall, we found that out of 38 predicted miR-124 canonical 3’UTR targets tested, 37 showed statistically significant downregulation at the protein level (p<0.01, t-test with Sidak correction for multiple testing, Figure 1C). Only 1 out of 38 (2.6%) tested miR-124 seed targets did not show significant downregulation, which is in good agreement with the 4.1% false discovery rate based on the calculated probability that a miR-124 seed site occurs in a Ciona 3’UTR by random chance. The percent downregulation of tested targets ranged from ~15% to ~90%. All 10 of the tested control non-targets showed no significant downregulation, although curiously one non-target showed a significant up-regulation, which was consistent in three independent replicates (Figure 1C). We previously showed that mutating the target site abrogated downregulation for all five tested seed knockout constructs and actually observed upregulation for three of these (Chen et al, 2011) (Table S1). There is evidence that saturating the RNAi machinery by overexpressing a certain miRNA construct can cause upregulation of gene targets for other miRNAs, and so we suspect that our observed upregulation of the one non-target and three site-mutated targets is due to saturation of the RNAi regulatory machinery by miR-124 overexpression (Khan et al, 2009). Incidentally, among our tested genes were four targets for which the canonical seed site was located in an alternatively spliced 3’UTR. Each of these targets (labeled ‘AS’ in Figure 1C) showed significant downregulation, whereas each of the corresponding non-target splice isoforms was not downregulated ( Figure 1C). This suggests that alternative splice isoforms with distinct 3’UTRs may exhibit differential miRNA regulation, and that expression of different 3’UTR isoforms may be a way to avoid or to promote miRNA regulation during specific developmental stages or in specific cell types (Mangone et al, 2010). |
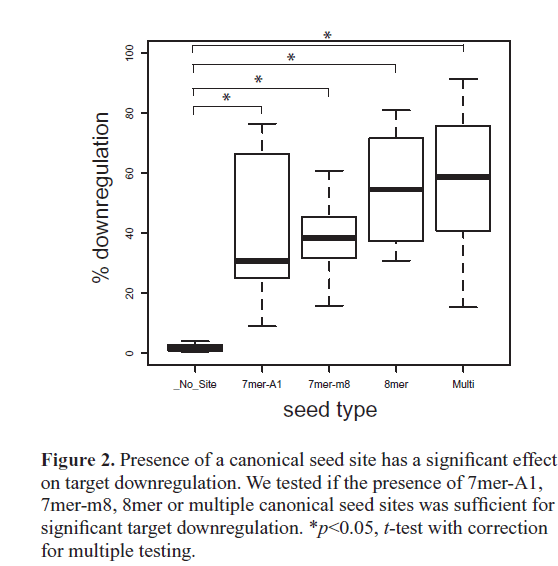
| On average, canonical targets containing multiple seed sites were the most downregulated, followed by 8mer, then 7mer-m8, and then 7mer-A1 being the least downregulated (Figure 2). This ordering of canonical seed classes in our in vivo assay agrees with previous in vitro high-throughput assays using both microarrays (Grimson et al, 2007) and SILAC proteomics (Baek et al, 2008; Selbach et al, 2008). Importantly, the average downregulation for each class of canonical seed targets was significantly higher than transcripts without a seed site, indicating that the presence of a 3’UTR canonical seed site has a significant effect on miR- 124-mediated gene knockdown (Figure 2). Our strongest target, Macho-, contains two seed sites and was downregulated 88.7%. We tested two additional single site constructs with one or the other seed site mutated, Macho-1-SeedKO1 and Macho-1-SeedKO2, and found that each single site construct was downregulated 84.9% and 77.9%, respectively. |
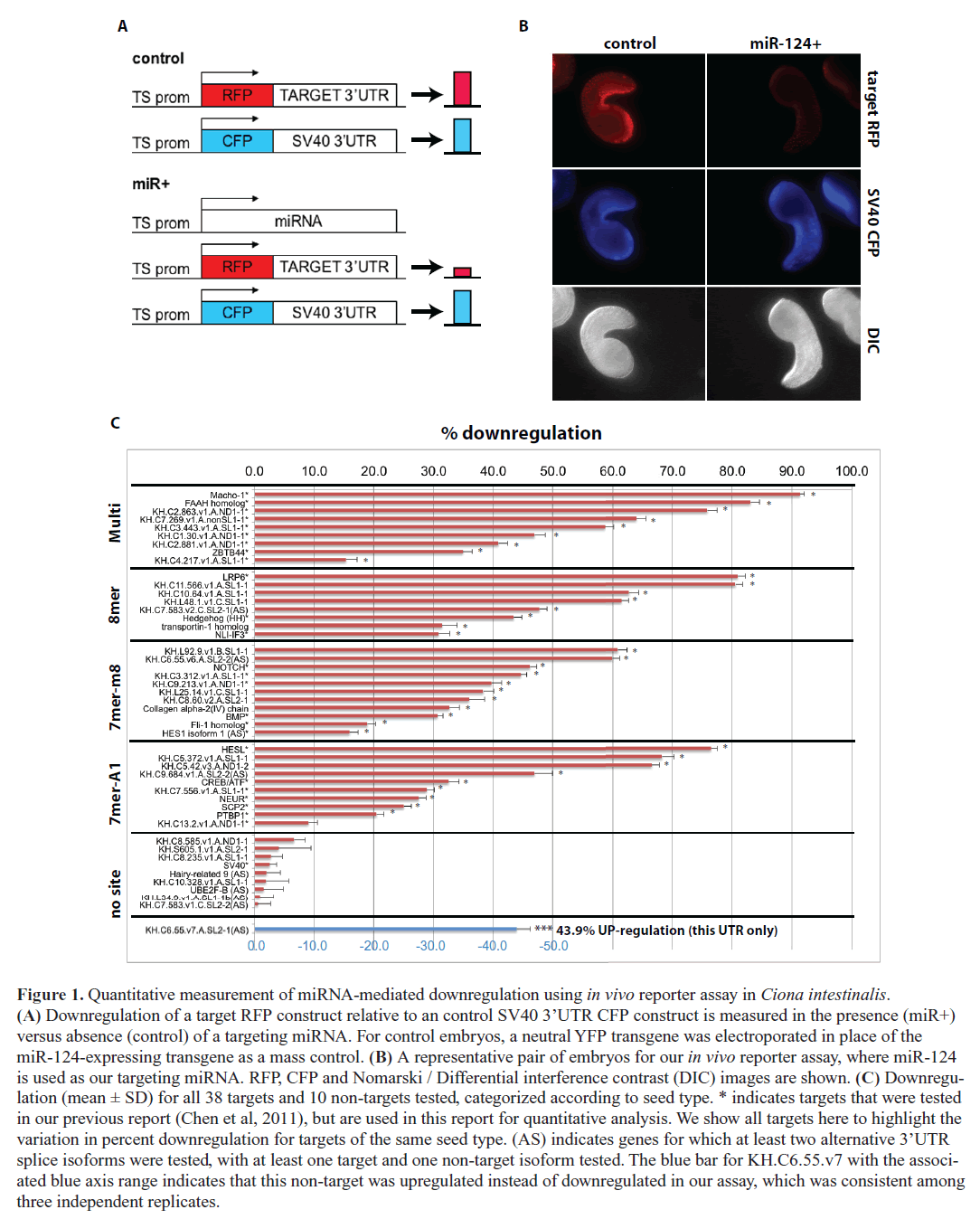 |
| This suggests a small degree of cooperative regulation between the two target sites, which are spaced ~80 nucleotides apart (Grimson et al, 2007; Saetrom et al, 2007). Simultaneously mutating both seed sites completely abolished Macho-1 downregulation (Table S1). In agreement with the previous observation that miR-124* and moR-124 expression levels are low during Ciona development (Shi et al, 2009), we observed that the genes we tested containing predicted miR-124* sites (KH.C8.585.v1) or moR-124 sites (KH.S605.1.v1, KH.C10.328.v1) in their 3’UTRs did not exhibit significant downregulation (Figure 1C). |
| Within each canonical seed class, we found a significant amount of variation in protein downregulation levels ( Figure 1C). For example, among 8mer targets we noticed a ~50% range in downregulation levels. Each of the targets was expressed at the same concentration and under the control of the same epidermal promoter, so differences in downregulation levels were not due to differences in expression level or timing. This suggests that there are determinants other than seed type that modulate the level of target downregulation. |
 |
| Correlation analysis of previously identified factors with miRNA-mediated target translational repression |
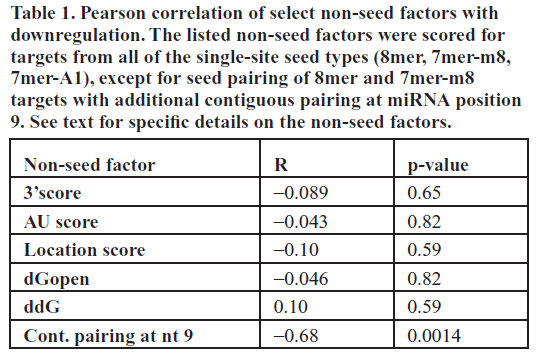
| Previous reports have suggested several other non-seed determinants that can affect miRNA target downregulation (Grimson et al, 2007; Kertesz et al, 2007; Nielsen et al, 2007; Bartel, 2009). To quantitatively investigate the effect of previously suggested factors modulating miRNA target downregulation at the protein level, for each single site miR-124 canonical seed target we determined the (1) site accessibility score ΔGopen, (2) free energy score ΔΔG, (3) AU-context score, (4) location score, (5) 3’end complementarity score, (6) the presence of an adenine or uracil opposite miRNA position 9 and (7) individual base pairing for all non-seed nucleotides (positions 9–22) of miR- 124 (Table S1). These are the major determinants that have been proposed to affect downregulation levels (Grimson et al, 2007; Kertesz et al, 2007; Nielsen et al, 2007; Bartel, 2009). Criteria (1) to (3) are related to the secondary structure surrounding the target site, while criteria (5) to (7) are related to pairing outside the seed. We determined non-seed pairing based on the miR-124:target duplex structure generated using RNAhybrid (Krüger and Rehmsmeier, 2006). Our tested targets spanned a wide range of scores for each of the criteria, allowing us to determine the degree of correlation between each factor and percent downregulation in our assay. We searched for overall correlation for single-site targets as well as correlation within each seed class. |
| Of all the factors analyzed, the only significant effect on target protein downregulation we observed in our assay was contiguous pairing up to nucleotide 9 for 8mer and 7mer- m8 canonical targets (R=-0.68, p=0.0014, Table 1). Perhaps contrary to the intuition that more non-seed pairing confers greater knockdown efficacy, additional base pairing at miRNA position 9 for these targets actually significantly decreased the average downregulation by ~30%. Although the mechanism for this decrease in knockdown efficiency is unknown, we do note that two previous studies, one using miRNA sponges and the other using anti-miR lentiviral vectors, found that introducing a bulge opposite miRNA positions 9–12 causes greater miRNA knockdown, presumably due to more stable interaction with the miRNA (reviewed in Ebert et al, 2010). None of the other previously identified determinants were independently correlated with the degree of target protein downregulation in our in vivo assay (t-test, Table 1). We also tested all multiplicative combinations of secondary structure [criteria (1) to (3)] and sequence [criteria (5) to (7)] factors, and also found that none of these joint effects were significantly correlated with downregulation in our assay (data not shown). |
 |
| Finally, recent studies have suggested two new classes of miRNA targets – those containing G-bulge seed sites (Chi et al, 2012) or imperfect centered sites (Martin et al, 2014). Since these sites may also mediate miRNA downregulation, we searched for G-bulge and imperfect centered sites among our tested miR-124 targets. In scanning all of our 38 tested targets, we found only 1 target containing a G-bulge site, but 22 targets contained at least one imperfect centered site (Table S1). However, the number of imperfect centered sites was not correlated with target downregulation levels (R=−0.10, p=0.55), and so the presence of these sites does not explain the variation in downregulation among our targets. |
| MicroRNA target regulation activity obeys Michaelis- Menten kinetics |
| It may be argued that the lack of correlation of non-seed determinants with downregulation may be an artifact of our sensor assay. One possibility is that perhaps overexpression of the miRNA vector at extremely high cellular concentrations saturates the endogenous silencing machinery (Borel et al, 2011), which may override any potential effects of other determinants such as secondary structure. Previous studies have demonstrated that the miRNA machinery can become saturated, thus affecting the ability of miRNAs to properly regulate their target genes (Khan et al, 2011; Grimm, 2011). In addition, it is unclear how changes in relative miRNA concentration levels affect target downregulation. |
| To address these questions, we selected seven representative targets spanning the different seed classes and repeated our in vivo sensor assays with titrations of our miR-124 expressing transgene in the range of 1/8X to 2X the molar concentration relative to the sensor target. For Macho-1, we also performed titration assays for the two seed knockout constructs, Macho1-SeedKO1 and Macho1-SeedKO2. |
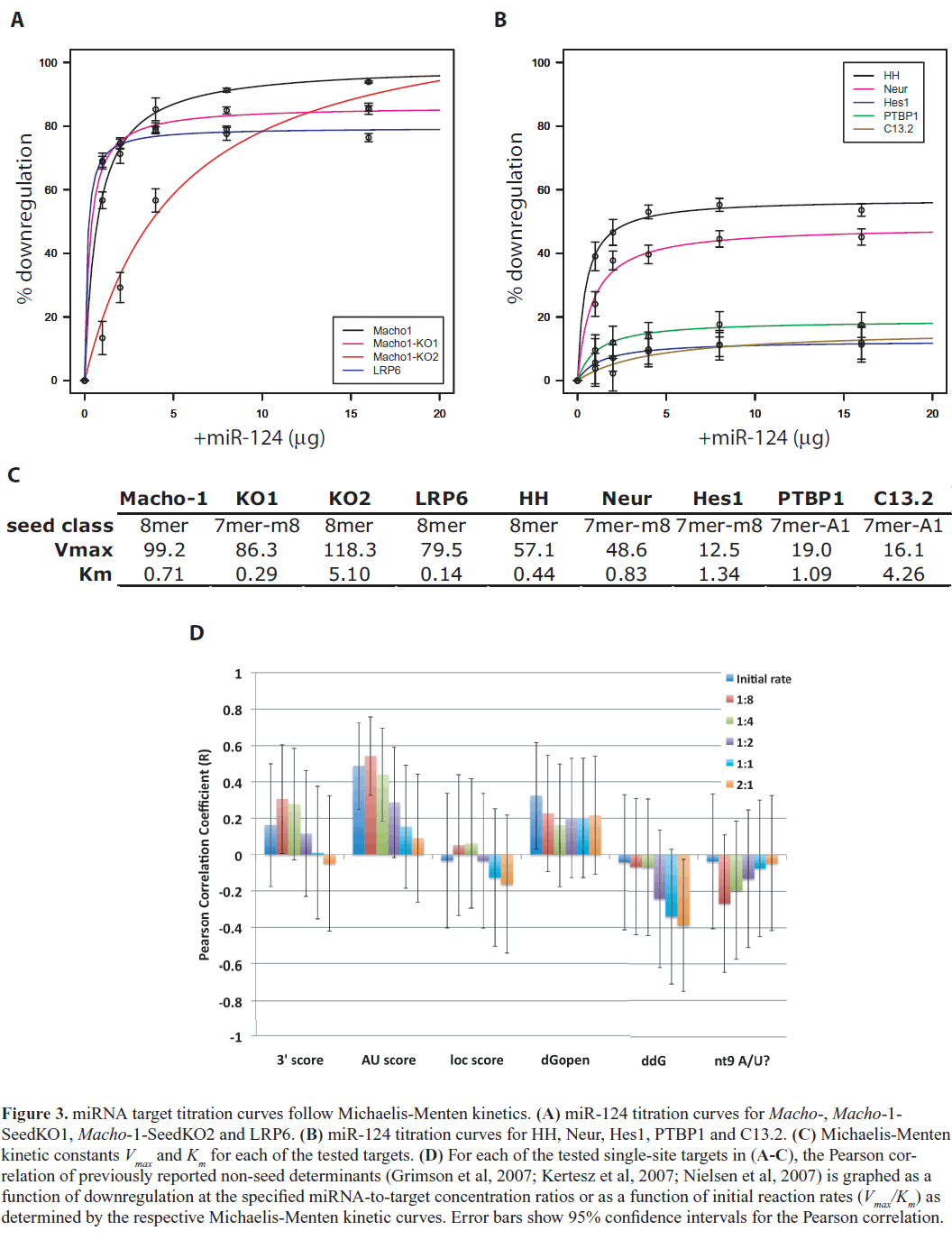 |
| Figure 3 shows the results of our titration assays for each of these targets. We observed that downregulation levels appear to saturate at high miR-124 concentrations – i.e., there is a maximum downregulation for each target. The 1:1 concentration ratio used in our initial assays was already nearsaturation. Interestingly, we found that the titration curves for each of our tested targets closely followed Michaelis Menten kinetics, suggesting that the kinetics of interaction of a miRNA with its binding site resembles the interaction between an enzyme and its substrate, and mathematically may be modeled accordingly. In other words, the regulatory activity of the Argonaute/RISC complex on a miRNAtarget duplex behaves according to Michaelis-Menten enzyme kinetics. The regulatory activity (i.e., downregulation) approaches a maximum threshold Vmax, representing saturation of the Argonaute/RISC machinery. Compared to Michaelis-Menten kinetics, other types of dose-response models such as linear, logistic and exponential models did not fit our titration curves as well (Table S2). Our model is consistent with a previous study that showed initial transcript degradation profiles after miRNA transfection in cell culture fit Michaelis-Menten kinetics better than linear or constant models (Arvey et al, 2010). The dissociation constant, Km, was of the same order of magnitude and fairly consistent between all targets, suggesting a general, target-independent process of loading of the target RNA substrate into the active miRISC complex (Figure 3C). Notably, there was a fairly high degree of correlation between the initial reaction rate and percent downregulation (Pearson correlation coefficient, R=0.63). This correlation is significant when we exclude Macho-1-SeedKO2 (R=0.76, p=0.03), which was an experimental outlier with abnormal kinetic constants. Previous studies have shown that miRNA saturation generally modulates downregulation of all target genes (Arvey et al, 2010; Khan et al, 2009); here we show that the level of downregulation at saturation levels varies among individual targets. |
| Since different targets have different degrees of maximum downregulation, this suggests that there are sequence and/or structural determinants that affect the efficiency of miRISC activity. Although our analysis showed that the non-seed determinants are not correlated with maximum downregulation, through analyzing our titration data we observed that among the non-seed determinants, two factors related to 3’end complementarity (3’score and nt9 A/U) and a factor related to secondary structure (AU score) were increasingly more correlated with downregulation at lower miRNA concentrations (Figure 3D). Correlation of downregulation with initial reaction rates was also much higher for AU score and 3’score (Figure 3D). This suggests that, while previously identified non-seed determinants are not correlated with maximal levels of downregulation and may not affect the efficiency of miRISC regulation, secondary structure and 3’end complementarity determinants may play a role in the initial miRNA-target binding interaction and miRISC activity at lower concentrations. |
CONCLUSIONS |
| Using a previously developed in vivo sensor assay to test and quantify downregulation of natural canonical miRNA targets in Ciona intestinalis, we were able to verify that increased seed base pairing and increased number of seed sites in a target transcript result in higher levels of downregulation at the protein level. We also found that the target response is dose-dependent and obeys Michaelis-Menten kinetics, suggesting that miRISC-target interactions resemble enzyme-substrate interactions. Furthermore, target downregulation correlates with the initial reaction rates and saturates at high miRNA concentrations, with each target saturating at different levels depending on its kinetic profile. Our results highlight the often-overlooked importance of relative cellular concentration levels of the miRNA and target when designing miRNA-based RNAi strategies and also for studying the mechanisms of endogenous miRNA regulation. Our data also hints at the importance of secondary structure and 3’end complementarity for target regulation upon initial miRISC-target binding and at low relative miRNA concentrations. Together, these results suggest that relative target expression levels and non-seed properties can explain why miRNAs can act as a regulatory ‘off ’ switch for a few targets and as a fine-tuner of gene expression for others. Our results indicate that there are still yet undiscovered non-seed factors that contribute to miRNA targeting. With recent advances in genome editing based on CRISPR technology (Doudna and Charpentier, 2014), another exciting possibility will be using endogenous editing of miRNA target sites to alter miRNA-mediated regulation. This possibility, as well as the rational design of miRNA-based RNAi therapies, will depend on the further elucidation of nonseed factors contributing to miRNA knockdown efficacy. Finally, it should be emphasized that our work was done in Ciona intestinalis, a chordate with a highly AT-rich genome that is the closest invertebrate relative to vertebrates. For more direct application to human biology, future work will need to use similar approaches for investigating non-seed factors in a mammalian organism, for which the rules of miRNA targeting may be slightly different. |
ACKNOWLEDGMENTS |
| This work was funded by the National Science Foundation (NSF) [grants IOS0347937, IOS0951347 to RWZ]. JSC thanks lab members V Hurless, F McQueen, M Whedbee and D Velasco for their help and advice throughout the project. |
COMPETING INTERESTS |
| None declared. |
References |
|