Research Article - Journal of Veterinary Medicine and Allied Science (2018) Volume 2, Issue 1
Morphological and molecular characterisation of common amphistome species from cattle of South India
Shameem H1*, Devada K1, Lakshmanan B1, Joseph S1, Sabu L2, Usha AP31Department of Veterinary Parasitology, College of Veterinary and Animal Sciences, Mannuthy, Kerala, India
2Department of Veterinary Microbiology, College of Veterinary and Animal Sciences, Pookode, Kerala, India
3Centre for Pig production and Research, Mannuthy, Kerala Veterinary and Animal Sciences University, Pookode, Kerala, India
- *Corresponding Author:
- Shameem H
Department of Veterinary Parasitology
College of Veterinary and Animal Sciences
Mannuthy
Kerala
India
Tel: +9847008341
E-mail: shameem@kvasu.ac.in
Accepted Date: February 24, 2018
Citation: Shameem H, Devada K, Lakshmanan B, et al. Morphological and molecular characterisation of common amphistome species from cattle of South India. J Vet Med Allied Sci 2018;2(1):7-11.
Abstract
Bovine amphistomosis is a highly neglected snail borne trematode disease causing great economic loss to dairy farmers. The aim of the present study was morphological identification of different amphistome species and molecular characterization of the most common amphistomes prevalent in the study area. Among the ten different species of amphistomes identified in the present study four common species namely, F. c o b b o l d i, F. e l o n g a t u s, G. c r u m e n if e r and Paramphistomum spp. were genetically characterized by amplifying second internal transcribed spacer sequence flanking 5.8S and 28S partial ribosomal gene sequences (ITS-2+) which produced amplicons of 494 bp, 503 bp, 514 bp and 494 bp respectively. The nucleotide sequences of four species of amphistomes F. cobboldi, F. elongatus, G. crumenifer and Paramphistomum spp . obtained in the present study is the first report from Kerala (India) which provides the primary sequence data of the rDNA ITS-2+ region of the amphistome flukes. Nucleotide sequence analysis of the four species revealed eight nucleotide differences between the sequences of pouched amphistomes ( F. cobboldi, F. elongatus and G. crumenifer ) and unpouched ( Paramphistome spp.). Nucleotide sequence analysis and in silico restriction map analysis of four amphistomes suggest the possibility of ITS-2+ region as a useful genetic marker for species identification.
Keywords
Amphistomes, Cattle, PCR, ITS2.
Introduction
Amphistomosis is a common ruminant trematode disease causing high morbidity and mortality in cattle, goats, sheep and water buffaloes. The disease is characterized by gastroenteritis, reduced milk production and lowered feed conversion resulting in economic loss. The disease has widespread geographical distribution in tropical and subtropical areas and has recently emerged as an important cause of productivity loss [1]. Paramphistomosis is caused by trematodes belonging to different genera and species of the family Paramphistomidae.
Even though these species are difficult to identify due to their morphological similarities they may differ in certain aspects like susceptible hosts, pathogenicity and geographical distribution. More than forty species of amphistomes have been recorded, among which four species viz., Gastrothylax crumenifer, Fischoederius cobboldi, F. elongatus and Paramphistomumi are regarded as common amphistomes of domestic ruminants [2]. Paramphistomes are ubiquitous and abundant within hosts but the importance of these worms has been underestimated widely.
Since amphistome species are morphologically similar alternative approaches using molecular biology techniques are useful for species identification [3,4]. The second internal transcribed spacer (ITS2) region of ribosomal DNA (rDNA) is particularly shown to be a useful genetic marker in species identification and for phylogenetic analyses [3,5-7].
Hussain et al. [7] characterized three different genus of amphistomes based on rDNA ITS2+ region whereas Ghatani et al. [8] derived molecular phylogeny of five gastrothylacid species using the ITS2 sequence and secondary structure analyses.
Little attention has been received in the use of molecular tools to identify the species prevalent in South India.
Molecular characterization of the prevalent species of our area will help to explore the regional variation and to contribute a molecular phylogenetic framework for paramphistomes in Kerala.
Apart from morphological identification the present study thus aimed to characterize the ITS2+region of four predominant amphistome species viz., Gastrothylax crumenifer, Fischoederius cobboldi, F. elongatus and Paramphistomumi spp. infecting ruminants in South India.
Materials and Methods
Collection of flukes
Adult amphistomes were collected from rumen of naturally infected cattle slaughtered at Municipal Corporation Slaughter house, Thrissur, Kerala. The parasites were washed thoroughly in phosphate buffered saline (PBS), pH 7.4 and identified by morphology as per Nath [9]. Individual flukes after washing were pressed gently in between two microscopic slides for gross identification of the species. The representative samples of different species of amphistomes were grouped into two for morphological and molecular studies.
Morphological identification
First group was processed using conventional techniques for species confirmation microscopically. Permanent microscopic slides were prepared using carmine stain as per standard procedure [10] and were used for detailed morphological studies.
Molecular studies
The second group which was morphologically identified as Gastrothylax crumenifer, Fischoederius cobboldi, F. elongatus and Paramphistomumi spp. were stored in -20C for DNA extraction.
DNA extraction was performed using the DNeasy blood and Tissue Kit (QIAGEN, Hilden, Germany) according to manufacturer’s recommendations with necessary modifications. Single adult fluke from each species was used for the extraction of genomic DNA. The DNA concentration at 260 nm and purity at 260:280 nm was determined using nanospectrophotometer (Nano drop 200C, Thermoscientific, USA).
The PCR was performed in 25 μl reaction using primers targeting second internal transcribed spacer plus flanking 5.8S and 28S (ITS-2+) region of ribosomal DNA (r DNA) as per using [8] published primer 3S (forward):5’- GGTACCGGTGGATCACTCGGCTCGGTG-3’and A28 (reverse):5’-GGGATCCTGGTTAGTTTCTTTTCCTCCGC-3’. A master mix was prepared without template containing 200μM dNTPs (Merck Genei, Banglore), 1.5 mM MgCl2, 25pMol of each primer, 1 unit Taq DNA Polymerase (Sigma Aldrich, Banglore) and 2.5 μl 10xPCR buffer.
PCR was performed in a thermocycler (MJ Mini, BioRad) under the following cycling conditions for different DNA templates (DNA from G. crumenifer, F. cobboldi, F. elongatus, Paramphistomumi spp.): 94C for 5 min; 35 cycles of 94C for 1 min, 75C for 30 sec and 72C for 1 min; followed by 72C for 7 min.
The amplified PCR products were identified in 1.5 per cent agarose gel prepared in Tris-borate EDTA (TBE) buffer containing Ethidium bromide (SRL, Mumbai), visualised and photographed in a Chemi DocTM MP Imaging System (Bio Rad, USA).
The amplicons were sent to Sci Genom Labs Pvt. Ltd., Kakkanad, Cochin for column purification and custom bidirectional sequencing using Sanger’s dideoxynucleotide chain termination method. The sequences were blasted using NCBI BLASTn tool for further confirmation and submitted to the GenBank through Banklt.
Nucleotide sequence analysis
Nucleotide Sequence alignment of individual amphistomes was carried out using CAP3 online software [11] and multiple sequence alignment was done using CLUSTAL O (1.2.1) software [12].
In silico analysis of the ITS-2+ sequences was performed to identify the restriction enzyme (RE) cutting sites using NEB cutter online tool (BioLabs).
Phylogenetic analysis was performed with DAMBE [13] online database software (http://dambe.bio.uottawa.ca/ dambe.asp) using corresponding gene sequences of ITS-2+rDNA gene sequences of related amphistomes available in the Genbank (www.ncbi.nlm.nih.gov).
Results
Morphological characterization of amphistomes
Adult amphistomes collected from rumen of cattle were subjected to morphological identification as described by Nath [9] (Plate 1).
Based on the shape and position of testes, uterus, oesophagus, intestinal caeca and genital sucker, ten different species of flukes were identified namely Fischoederius cobboldi, G. crumenifer, F. elongatus, Carmyerius spatiosus, P. epiclitum, P. cervi, Ceylonocotyle scoliocoelium, scoliocoelium cotylophorum, C. indicum and explanatum explanatum (Gigantocotyle explanatum) (9, 14, 15) (Plates 2-4).
Among the different pouched amphistomes identified F. cobboldi was the most prevalent species followed by G. crumenifer as revealed from slaughter house studies. Mixed infection was common with pouched amphistomes outnumbering unpouched ones. Every infected rumen carried thousands of flukes with suckers adhered to the ruminal papillae and mucosa.
Molecular characterization of amphistomes
Polymerase chain reaction was standardised with DNA extracted from the four common amphistomes namely F. cobboldi, F. elongatus, G. crumenifer and Paramphistomumi spp. using published primers amplifying second internal transcribed spacer (ITS-2) complete sequence and flanking 5.8S and 28S partial ribosomal gene sequences (ITS-2+).
The standardised protocol of PCR with an annealing temperature of 75°C amplified an approximately 500 bp product for all the four species namely F. cobboldi, F. elongatus, G. crumenifer and Paramphistomumi spp. using ITS-2+ primers.
The amplicons of four amphistomes namely F. cobboldi, F. elongatus, G. crumenifer and Paramphistomumi spp. obtained after PCR with ITS-2+ primers were of 494 bp, 503 bp, 514 bp and 494 bp, respectively (Figure 1).
The sequences were aligned using online software CAP3 and blasted using NCBI BLASTn tool to analyse their identity with other published sequences available in online databases. The sequence length of four different species of amphistomes obtained with ITS-2+ rDNA primers along with their accession numbers obtained are summarised in Table 1.
Table 1: Sequence length and accession numbers of amphistomes.
| Species | Sequence Length | Accession number |
|---|---|---|
| Fischoederius cobboldi | 494 bp | KU530202 |
| Gastrothylax crumenifer | 514 bp | KU530204 |
| Fischoederius elongatus | 503 bp | KU530203 |
| Paramphistomumi spp. | KU530205 |
Multiple sequence alignment of the ITS2 rDNA sequences of F. cobboldi, G. crumenifer, F. elongatus and Paramphistomumi spp. were carried out using CLUSTAL O (1.2.1) online software.
Nucleotide sequence analysis of the four species revealed eight nucleotide differences between the sequences of pouched amphistomes G. crumenifer, F. cobboldi, F. elongatus and the unpouched species (Paramphistomumi spp.).
The ITS-2+ sequences of the two Fischoederius spp. obtained in the present study were 100 per cent identical whereas it revealed nucleotide differences at two bases with the corresponding sequence of G. crumenifer (Figure 2).
In Silico restriction enzyme map analysis
In Silico Restriction Enzyme Mapping of the ITS-2+ rDNA sequences of F. cobboldi, F. elongatus, G. crumenifer and Paramphistomumi spp. was performed using the online software NEB cutter.
It revealed that the pouched amphistomes share several restriction enzyme recognition sites. The analysis of G. crumenifer sequence revealed unique recognition sites for BtsI, MutI and TspRI which cut the sequence at 330 bp position.
The sites for these enzymes were lacking in F. cobboldi, F. elongatus and Paramphistomumi spp. The sequence of Paramphistomumi spp. differed from the pouched species in the absence of recognition site for the enzyme BspCNI which could produce 335 bp and 179 bp products in the latter.
Phylogenetic analysis
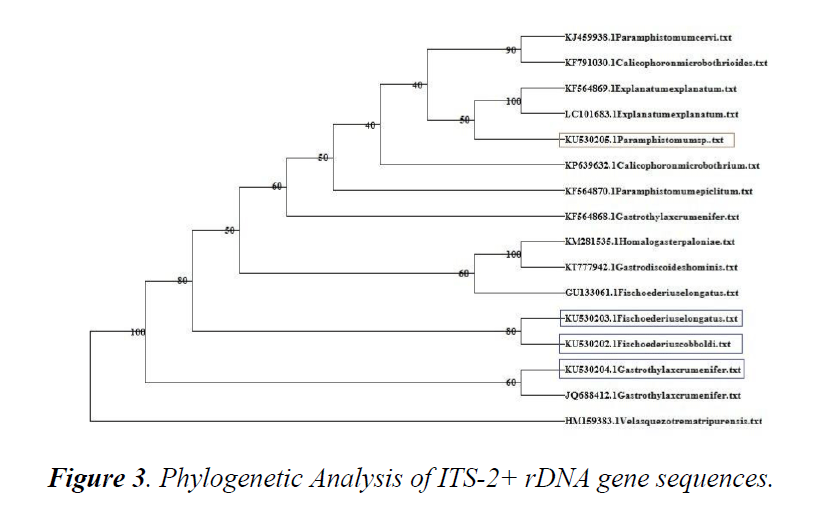
A phylogenetic tree using online software DAMBE was constructed using sequences available in online database.
Sequences were retrieved and aligned using CLUSTAL W with ITS2+ rDNA sequences of F. cobboldi, F. elongatus, G. crumenifer and Paramphistomumi spp. obtained in the present study. Based on phylogenetic analysis, G. crumenifer, Kerala shared the same clade with the same species isolated from Meghalaya (JQ688412.1). Kerala isolates of F. cobboldi, F. elongatus and Shillong isolate of F. elongatus (GU133061) clustered together.
The unpouched amphistome species occurred in a different clade from the pouched species. Paramphistomumi spp. Kerala isolate shared the same clade with the unpouched amphistome isolates from Bairelly and Japan (Figure 3).
Discussion
is even more difficult. Thus alternative approach to delineate species is needed. In the present study, ten different species of amphistomes were identified based on morphological parameters from abattoir samples viz., F. cobboldi, G. crumenifer, F. elongatus, Carmyerius spatiosus, P. epiclitum, P. cervi, Ceylonocotyle scoliocoelium, scoliocoelium cotylophorum, C. indicum and explanatum explanatum. Earlier work on the prevalent species of amphistomes by Nath [9] recorded seventeen species of amphistomes from Kerala amongst which G. crumenifer was predominant. However, in the present study F. cobboldi was found to be the most predominant species. The present study used DNA based tools for authentic identification of the common flukes recovered. PCR-based diagnosis using rDNA ITS-2 sequences have been proved as a reliable tool to identify digenean species and to recover their phylogenetic relationships [5]. The second internal transcribed spacer (ITS-2) sequence flanking 5.8S and 28S partial ribosomal gene sequences of the four species of amphistomes viz., F. cobboldi, F. elongatus, G. crumenifer and Paramphistomumi spp. were amplified using ITS-2 primers with sequenced product sizes of 494 bp, 503 bp, 514 bp and 494 bp for F. cobboldi, F. elongatus, G. crumenifer and Paramphistomumi spp. respectively. The second internal transcribed spacer region of rDNA is particularly valuable and can be utilized for phylogenetic analyses at the genus and species levels [3,14-17]. Similar sequencing studies were carried out by Hussain et al. [8] who reported amplification of an approximately 500 bp of the ITS-2+ region for P. epiclitum, G. crumenifer and G. explanatum. The present study is also in agreement with Ghatani et al. (2012) who recorded 477 bp, 520 bp, 463 bp and 482 bp products by amplifying ITS2+ region of F. cobboldi, F. elongatus, G. crumenifer, C. spatiosus and V. tripurensis. Shylla et al. [16] genetically characterised three species of trematodes, H. paloniae, C. calicophorum and O. streptocoelium using ITS-2 primers and obtained annotated products of 239 bp, 288 bp and 288 bp. ITS-2 sequence analysis revealed eight nucleotide differences between the sequences of pouched (F. cobboldi, F. elongatus, G. crumenifer) and unpouched (Paramphistomumi spp.) species. Besides it revealed 100 per cent identity between the two Fischoederius species but was clearly different from G. crumenifer at two nucleotide positions. Ichikawa et al. [16] reported that Gigantocotyle explanatum differed from P. leydeni at 7 nucleotide sites and hence could be used as a molecular marker for epidemiological studies on G. explanatum. Differences of even one nucleotide change in the ITS-2 sequence can be used as an effective genetic marker to distinguish closely related digenean species [8]. Hence in the present study the two nucleotide difference between G. crumenifer and Fischoederius species suggests the possibility of ITS-2+ region to be used as a species identification marker. However, further studies are required with more number of sequence data for confirmation. Moreover, the results of the study will contribute to the sequence data base of South Indian isolates of amphistome species. The present study reports sequence divergence upto 5.4 among the four amphistome species suggesting the usefulness of the ITS-2+ region for discriminating closely related species. Restriction mapping of the sequences obtained in the present study when analysed in silico revealed that the four amphistomes shared the restriction sites for a number of endonucleases. Gastrothylax crumenifer was found to have unique recognition site for BtsI MutI and TspRI which were absent in F. cobboldi, F. elongatus and Paramphistomumi spp. Also the present study identified the recognition site for the enzyme BspCNI only in pouched amphistomes which differed from unpouched Paramphistome spp. Thus restriction enzyme mapping in the present study suggests the possibility to differentiate G. crumenifer from the other species with the presence of recognition site for BtsI MutI and TspRI. Itagaki et al. [3] identified three amphistome species by PCR-RFLP based on the differences in the restriction sites of AccII on the ITS2 sequence. Phylogenetic tree topology obtained based on the sequence data suggested that the four amphistome species belong to two families viz., Gastrothylacidae and Paramphistomidae. The Kerala isolates obtained in the present study seemed to cluster with Meghalaya and Shillong counterparts. Ghatani et al. [7] had shown the applicability of ITS-2 sequence and secondary structure in deriving phylogenetic relationships at interfamily level. Hussain et al. [8] reported sequence homology in isolates of P. epiclitum (99.3-99.7%), G. crumenifer (97.2-99.3%) and G. explanatum (95.1-97.2%) on phylogenetic analysis. The present study reports the first molecular characterisation of amphistome flukes from the state of Kerala, India. This study confirms ITS2 as a useful molecular marker for species identification and the sequence data obtained serves as a corner stone for further epidemiological studies on amphistomes in the region.
Acknowledgement
The authors are thankful to the Kerala Veterinary and Animal Sciences University for the financial grant and facilities provided for the research work.
References
- Huang J, Jia R, Wang M, et al. An Attenuated Duck Plague Virus (DPV) Vaccine Induces both Systemic and Mucosal Immune Responses To Protect Ducks against Virulent DPV Infection. Clinical and Vaccine Immunology. 21;4:457-62.
- Kulkarni DD, James PC, Sulochana S. Assessment of the immune response to duck plague vaccinations. Research in Veterinary Science. 1998;64:199-204.
- Stallknecht DE, Hunter DB, Slemons RD. Avian Influenza. Infectious diseases of wild birds edited by Nancy J. Thomas, D. Bruce Hunter, Carter T. Atkinson 2007, pp: 108-30.
- Kayali G, Kandeil A, Rubrum A, et al. Active surveillance for avian influenza virus. Egypt, 2010-2012. Emerg Infect Dis. 2014;20:542-51.
- Allan, Lancaster WH, Toth B, et al. Newcastle disease vaccines: their production and use. FAO. Animal production and health series No.10.FAO. Rome. Indian J Anim Sci. 1978;61:357-9.
- Lennete EH. Diagnostic procedures for viral and rickettsia diseases, (3rd edn.) A public health Ass.Inc.; Broadway 1964.
- Anon Y. Methods for Examining Poultry Biologics and for Identification and Quantifying Avian Pathogens. National Academy of Science, Washington DC, USA. 1971; 66.
- Bass EP, Gill MA, Beckenhaur WH. Development of a modified live canine origin parvo virus vaccine. J Am Vet Med Assoc. 1982;181:909-13.
- Singh KV, Osman OA, Thaana I, et al. Colostral transfer rinderpest neutralizing antibodies to offspring of vaccinated dams. Can J Comp Med Vet Sci. 1967;31:295-8.
- Voller A, Bartlett A, Bidwell DE, et al. The detection of viruses by enzyme-linked immunosorbent assay (ELISA). J Gen Virol. 1976; 33:165-7.
- Lin W, Lam KM, Clark WE. Active and passive immunization of ducks against duck viral enteritis. Avian Diseases. 1984;28:968-73.
- Swayne DE. Avian influenza. John Wiley & Sons. 2009.
- Tian G, Zhang S, Li Y, et al. Protective efficacy in chickens, geese and ducks of an H5N1inactivated vaccine developed by reverse genetics. Vaccine. 2005;341:153-62.
- Kumar M, Chu HJ, Rodenberg J, et al. Association of serologic and protective responses of avian influenza vaccines in chickens. Avian Diseases. 2007;51:481-3.
- Anhar A, Hayam F, Nermein Mahmoud, et al. Evaluation of duck immune response to mutual vaccination with avian influenza and duck hepatitis vaccines. SCVMJ, XV 2010;1: 245-54.
- Farouk H, Mahmoud N, Susan S. Comparative immunological studies between tissue culture and egg adapted duck plague vaccines. Report and Opinion. 2012;4:12.
- Hanan AA. Laboratory assessment of protection given by experimental Pasteurella multocida vaccines in ducks. Fac Vet Med. 2004.
- Dung DV, Loi DT, Meers J. Laboratory trials of a new duck plague vaccine produced in chicken embryo fibroblast cell cultures, Control of Newcastle disease and duck plague in village poultry. 2004; 59.