Research Article - Biomedical Research (2017) Volume 28, Issue 11
MiR-1291 may involve renal up-regulation of urate transporter 1 in patients with uric acid nephrolithiasis
Weihua Fu1,2#, Qianwei Li2#,Jiwei Yao2, Haoyu Wang2, Guoxian Deng2, Linyong Dai2, Junan Yan2* and Weibing Li2*
1Department of Urology, Xinqiao Hospital, Third Military Medical University, Chongqing, PR China
2Department of Urology, Southwest Hospital, Third Military Medical University, Chongqing, PR China
#These authors contributed equally to this work
- *Corresponding Authors:
- Junan Yan
Department of Urology
Southwest Hospital
Third Military Medical University, PR China
Weibing Li
Department of Urology
Southwest Hospital
Third Military Medical University, PR China
Accepted date: March 25, 2017
Abstract
Uric Acid (UA) nephrolithiasis is a constant threat to worldwide public health, its underlying pathogenesis is not elucidated yet. We previously identified that the overexpression of Urate Transporter 1 (URAT1) in the kidney was closely related with the UA nephrolithiasis, but the up-regulation mechanism was unclear. The aim of this study was to identify the possible miRNAs involved in regulating the renal expression of URAT1 in UA Stone Formers (UASF). MiRNAs microarray analysis revealed seven down-regulated miRNAs in the renal cortex of UASF compared with normal controls. Bioinformatic analysis suggested miR-1291, only one of the candidate miRNAs, could bind the 3'UTR of URAT1 gene. Furthermore, the down-regulated MiR-1291 and overexpression of URAT1 in the kidney of UASF were detected with real-time PCR. Thus, we inferred that the URAT1 gene was modulated by miR-1291, which was involved in up-regulated renal expression of URAT1 in UASF. More persuasive conclusion needs further research.
Keywords
miR-1291, Urate transporter 1, Uric acid nephrolithiasis
Introduction
Uric Acid (UA) nephrolithiasis constantly accounts for 5-10% of all urinary calculi [1]. The underlying pathophysiology of UA nephrolithiasis is complex and unclear, despite much investigation on the subject. In our previous study, significantly higher expression of Urate transporter 1 (URAT1, encoded by SLC22A12) was found in the renal cortex of UA Stone Formers (UASF) compared to normal controls, which might be involved in the development of UA stones [2]. However, the mechanism of the up-regulated expression of URAT1 is still unclear and need further research.
MicroRNAs (miRNAs) as a large family of highly conserved non-coding RNA consist of approximately 22 endogenous nucleotides. They can modulate target gene expression by inhibiting translation or inducing mRNA degradation at the post-transcriptional level, result in preventing target protein synthesis. MiRNAs play important roles in multiple physiological and pathological processes [3]. For example, miR-9 and miR-374 can significantly inhibited the gene expression of CLDN14 in the thick ascending limb of the human kidney, which is a key regulator for renal Ca2+ homeostasis [4]. The synonymous variant in CLDN14 gene is associated with kidney stones, which may due to CLDN14 escaping from miRNAs suppression [5]. A recent report showed that miR-34a could inhibit the gene expression of SLC22A12 in HEK293 cells [6]. However, it is still not known whether miRNAs are involved in regulation of SLC22A12 expression in the renal tissue. The aim of this study was to investigate the possible miRNAs targeting SLC22A12 in patients with UA nephrolithiasis.
Materials and Methods
Study participants
The study was approved by the ethics committee of Southwest Hospital Affiliated to Third Military Medical University. All participants were adult male and provided informed consent voluntarily. A total of 13 pairs of renal specimens were collected from patients in this study: patients with UA nephrolithiasis (group I) and normal controls (group II), who were recruited from inpatients of our center of urology from December 2013 to February 2015. UASF underwent Percutaneous Nephrolithotomy (PCNL) for stone removal. The renal samples of UASF were collected through percutaneous renal biopsy under ultrasound guided during the PCNL. Patients with recurrent urinary stones were excluded. Normal samples were obtained from patients who received emergency partial or radical nephrectomy owing to acute renal trauma, and who were healthy before accidents. The removed renal cortex was regarded as normal controls. Subjects were excluded when who were accompanied by documented familial inherited, gastrointestinal, liver or other renal disorders. Moreover, all participants did not receive any medications that may affect UA metabolism before this study. The informed consent was obtained from the College Ethics Committee.
Isolation and quality control of RNA samples
Total RNAs were extracted from samples of renal cortex using Trizol reagent (Life Technologies, USA) following the manufacturer’s protocols. In order to determine the concentration and quality of isolated RNA samples, total RNAs more than 5 μg of total RNAs were measured by the NanoDrop 2000 spectrophotometer (Thermo Scientific, USA) at 230 nm, 260 nm, 280 nm. The light source was Xenon flash lamp. And the operating power consumption was 12-18 W. Samples were required to meet the criteria of OD260/280>1.6 and OD260/230>1.0 [7].
MiRNA microarray analysis
In order to screen the MiRNAs of differential expression between samples of two groups, equal amounts of RNA samples from UASF and normal groups (n=9, respectively) were subjected to microarray analysis using the Human miRNA OneArray v5 (Phalanx Biotech, Hsinchu, Taiwan). Data were analysed with Rosetta Resolver System software (Rosetta Biosoftware, USA). Significantly differentially expressed miRNAs (intensity difference between two samples ≥ 1000, individual intensity ≥ 500, absolute Log2 fold change ≥ 1, and false discovery rate<0.05) were identified between the case-control groups, in which the down-regulated miRNAs were recognized as the candidate miRNAs.
Bioinformatic analysis
To reveal potential interactions between the candidate miRNAs and URAT1, bioinformatic analysis was performed via internet databases: TargetScan v5.1 (http://www.targetscan.org), miRanda (http://www.microrna.org/microrna/home.do), PicTar (http://pictar.mdc-berlin.de). The 3’ UTR sequences of URAT1 were examined to determine whether selected microRNAs showed “seed” matches (complementary at positions 2-8) and the minimum free energy was lower than 20 kcal/mol by using RNAhybrid application (http://bibiserv.techfak.unibielefeld. de/rnahybrid/). We only considered as potential miRNAs those who were all found match with URAT1 by three of these algorithms.
Quantitative real-time PCR
The extracted RNA of renal cortex was reversely transcribed using M-MLV-RTase (Promega, Madison, WI), according to the manufacturer’s instructions. The resulting cDNA was used for real-time PCR using the Applied Biosystems SYBR Green Master PCR Mix (Thermo Fisher Scientific) on the TP800 qPCR System (Takara Bio, Otsu, Japan). To normalize the data for quantification of URAT1 mRNA and the miR-1291, we used Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) and U6, respectively. The 2-ΔCT was used as the relative quantitation value of target genes to calculate the fold change. The primers sequences used in real-time PCR were as follows: URAT1, 5’-CCC CAC ACC CGT CCA CCT G-3’ (forward) and 5’-GGG CTG GGT TTG GGG GTT AAG-3’ (reverse); miR-1291, 5’-ACA CTC CAG CTG GGT GGC CCT GAC TGA AGA CC-3’ (forward) and 5’-TGG TGT CGT GGA GTC G-3’ (reverse); GAPDH, 5’-TGA CTT CAA CAG CGA CAC CCA-3’ (forward) and 5’-GGA GTG TTG GAG AAG TCA TAT TAC-3’ (reverse); U6, 5’-CTC GCT TCG GCA GCA CAT AT-3’ (forward); and 5’-TTG CGT GTC ATC CTT GCG-3’ (reverse). Each experiment was performed in triplicate and repeated three times.
Statistical analysis
All data were expressed as the mean ± SD. Statistical calculations were performed with SPSS 16.0 software (IBM, Armonk, NY). The Student t test was used to evaluate the differences between groups. Statistical significance was accepted as P<0.05.
Results
Clinical characteristics of participants
This study included 13 patients with UA stones and 13 normal controls. Clinical characteristics of the participants are summarized in Table 1. The demographic data suggested the mean age and Body Mass Index (BMI) were significantly higher in group I than group II (p<0.05), moreover, hypertension, hyperuricemia and diabetes were more prevalent in UASF compared with normal group. There were no significant differences in the incidence of BMI. According to the results of blood biochemical and urinalysis, higher serum concentrations of creatinine and UA, lower urine PH were observed in UASF group than normal controls (p<0.05).
| Variable | Group I (N=13) | Group II (N=13) |
|---|---|---|
| Demographic data | ||
| Age (y) | 50.31 ± 6.64? | 43.08 ± 7.46 |
| BMI (kg/m2) | 25.68 ± 5.03 | 23.61 ± 4.55 |
| Diabetes n (%) | 4 (30.8) ? | 0 |
| Hyperuricemia n (%) | 8 (61.5)? | 2 (15.4) |
| Hypertension n (%) | 2 (15.4)? | 1 (7.7) |
| Clinical parameters | ||
| Serum creatinine (umol/L) | 76 ± 21 | 69 ± 19 |
| Serum uric acid (μmol/d) | 477 ± 94? | 290 ± 71 |
| Serum calcium (mmol/L) | 2.19 ± 0.22 | 2.27 ± 0.25 |
| Urine pH | 5.61 ± 0.43? | 6.57 ± 0.46 |
Table 1. Clinical characteristics of the participants in this study.
MiRNAs of differential expression in the renal tissue of UASF
MiRNAs expression profiles were analysed through comparing between the UAFS and normal groups. The differential miRNAs were shown in Table 2. Five up-regulated miRNAs and eleven down-regulated miRNAs were identified in renal tissue of UASF compared with normal controls. Because upregulation of URAT1 expression was observed in the renal tissue of UASF in our previous study, we hypothesized that URAT1 expression may be at least partly regulated by miRNAs, therefore, the down-regulated miRNAs were identified as recognized as the candidates.
| Systematic name | Fold change (log2) | P value |
|---|---|---|
| Up-regulated | ||
| hsa-miR-3178 | 1.007638 | 0.037762 |
| hsa-miR-4792 | 1.001515 | 0.007695 |
| hsa-miR-5001-5p | 1.011976 | 0.046798 |
| hsa-miR-4539 | 1.10463 | 0.023836 |
| hsa-miR-4481 | 1.255189 | 0.004011 |
| Down-regulated | ||
| hsa-miR-10a-5p | -1.07322 | 0.036769 |
| hsa-miR-125a-3p | -1.02711 | 0.021644 |
| hsa-miR-126-3p | -1.13046 | 0.046756 |
| hsa-miR-1291 | -1.92469 | 0.017088 |
| hsa-miR-194-5p | -2.30172 | 0.012721 |
| hsa-miR-200a-3p | -1.27005 | 0.00763 |
| hsa-miR-29b-3p | -1.44356 | 0.006351 |
| hsa-miR-30a-5p | -1.82887 | 0.002699 |
| hsa-miR-3607-5p | -1.41846 | 0.02697 |
| hsa-miR-4284 | -1.75396 | 0.024658 |
| hsa-miR-5195-5p | -1.29395 | 0.005093 |
Table 2. Differential miRNAs in UASF vs. normal controls.
Prediction of the promising miRNAs regulating the expression of SLC22A12 gene
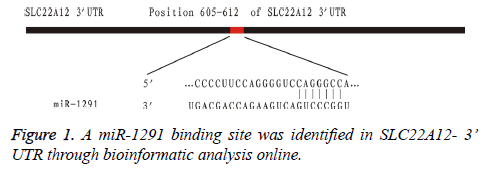
In order to identify the promising miRNAs regulating their target gene SLC22A12, the 11 down-regulated miRNAs were subsequently tested through bioinformatic analysis online. Among them, we found that miR-1291 was the only promising miRNA regulating the gene expression of SLC22A12. The possible target gene locus was identified between the miR-1291 and the sequences of SLC22A12 3’ UTR (Figure 1).
Gene expression of miR-1291 and SLC22A12 in renal tissues
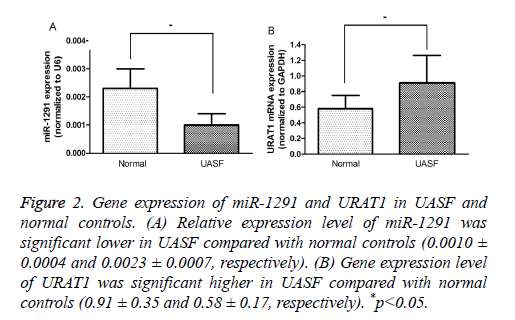
To confirm the result of prediction, the expression levels of miR-1291 and SLC22A12 were measured in the clinical samples by quantitative real-time PCR. The results suggested that the expression of miR-1291 was significantly lower in UAFS specimens than in the normal controls, instead, SLC22A12 was expressed significantly higher in renal samples of UAFS (P<0.05, Figure 2). Therefore, we believed that miR-1291 as a promising candidate was involved in regulation of SLC22A12 expression in the renal tissue.
Figure 2: Gene expression of miR-1291 and URAT1 in UASF and normal controls. (A) Relative expression level of miR-1291 was significant lower in UASF compared with normal controls (0.0010 ± 0.0004 and 0.0023 ± 0.0007, respectively). (B) Gene expression level of URAT1 was significant higher in UASF compared with normal controls (0.91 ± 0.35 and 0.58 ± 0.17, respectively). *p<0.05.
Discussion
The pathogenetic mechanisms for UA nephrolithiasis are diverse. According to the etiologies, it may be roughly divided into idiopathic, acquired, or idiopathic. Three urinary disorders have been always considered as the main factors for the development of UA nephrolithiasis: low urinary pH, hyperuricosuria and low urinary volume [8]. In the previous clinical studies, the relationship between UA nephrolithiasis and acidic urine has been recognized [2,9,10]. In addition, the epidemiologic studies revealed that UA nephrolithiasis was closely related to the metabolic syndrome (obesity and type 2 diabetes mellitus) [11,12]. Low urinary pH resulting from insulin resistance is also recognized as a significant role in its pathophysiologic mechanisms [13]. In this study, we also found the high prevalence of obesity or diabetes in patients of UA stones. However, the other urinary abnormalities, hyperuricosuria and low urinary volume, were not identified through 24 h urinalysis of UAFS in the clinical investigations, despite the higher plasma UA concentration [2,9,10]. The pathomechanism of the unexpected urate “low-excretion” is not elucidated currently. Our previous study suggested that it may be association with up-regulated expression of URAT1 in the kidney [2].
UA is the end production of purine degradation in humans, which can scavenge the potentially harmful reactive oxygen species, but hyperuricemia or hyperuricosuria may lead to UA nephrolitiasis. The homeostasis of blood-urine UA levers is mainly regulated by the urate transporting proteins in the human kidney. URAT1 play a key role in the renal urate handling, which is encoded by SLC22A12 and located in the proximal tubular epithelial cells. Plasmic UA can be freely filtered in the glomerulus, about 90% of filtered urate is subsequently reabsorpted by URAT1 [14]. In the past decade, much evidence deriving from genome-wide association studies suggested that the mutations or polymorphisms of SLC22A12 gene were closely associated with disorders of the urate metabolism, such as renal hypouricemia and hyperuricosuria, which could promote the formation of UA nephrolithiasis [15,16]. But as was mentioned above, excessive renal UA excretion or hyperuricosuria was not found in the clinical studies of UASF. Negri et al. [17] thought that impairment in renal urate excretion associated with increased serum UA was also a characteristic of UASF, which resembled patients with primary gout, but the underlying pathogenesis is still unclear. Our previous study found higher expression of URAT1 in the renal cortex of UASF compared with patients with calcium oxalate stones or normal controls, which might been involved in the abnormality of renal urate handling [2]. Doshi et al. [18] also reported that the cause of MetS-associated hyperuricemia appears to be associated with increased URAT1 protein in the mice kidney. However, the molecular mechanism for upregulated URAT1 is not still elucidated.
MiRNAs attract extensive attention due to their ability to negatively regulate about 30% of all protein-coding genes. Recently, Sun et al. reported that miR-34a could inhibit the gene expression of SLC22A12 in HEK293 cells [6]. In this study, five down-regulated miRNAs were indentified in renal cortex of UASF compared with normal controls by miRNA microarray analysis. We subsequently predicted the target genes of the candidate miRNAs through bioinformatic analysis and found that miR-1291 could bind the 3'UTR of SLC22A12 gene. Furthermore, expression of miR-1291 and URAT1 in the renal cortex of UASF were detected with real-time PCR, the results showed their negative correlation. The above results suggested that miR-1291 might regulate the expression of SLC22A12 gene in the renal cortex, the down-regulation of miR-1291 was involved in the over-expression of URAT1 in the kidney of UASF. Unfortunately, the further experiments was interrupted due to insufficient financial support, we could not provide more strong evidence to identify that URAT1 protein expression was regulated by miR-1291 in renal cells.
Conclusions
Altogether, this study showed the clinical characteristics of patients with UA nephrolithiasis. High plasma UA concentration, low urinary PH and relatively low-excretion of urate by the kidney were associated with UASF. In our previous study, up-regulated protein expression of URAT1 was thought to contribute to the abnormality of renal urate handling in UASF. According to the present evidence, we inferred that the down-regulation of miR-1291 might be involved in regulating the renal expression of URAT1 in UASF. More persuasive conclusion needs further research.
Conflict of Interest
None declared.
Acknowledgement
We thank Urology of Emergency Medical Center of Chongqing, China for providing renal samples. This study was supported by t the Natural Science Foundation of Chongqing Science and Technology Commission (Chongqing, China. Grant No. 2011BA5011).
References
- Shoag J, Tasian GE, Goldfarb DS, Eisner BH. The new epidemiology of nephrolithiasis. Adv Chronic Kidney Dis 2015; 22: 273-278.
- Fu W, Li Q, Yao J, Zheng J, Lang L. Protein expression of urate transporters in renal tissue of patients with uric acid nephrolithiasis. Cell Biochem Biophys 2014; 70: 449-454.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281-297.
- Gong Y, Renigunta V, Himmerkus N, Zhang J, Renigunta A. Claudin-14 regulates renal Ca2+ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J 2012; 31: 1999-2012.
- Thorleifsson G, Holm H, Edvardsson V, Walters GB, Styrkarsdottir U. Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet 2009; 41: 926-930.
- Sun WF, Zhang XX, Sun FY, Xu W, Liang J. MicroRNA expression patterns of the kidney in hyperuricemia mice treated with Xiezhuo Chubi Decoction. Chin J Integr Med 2011; 17: 35-42.
- Glasel JA. Validity of nucleic acid purities monitored by 260nm/280nm absorbance ratios. Biotechniques 1995; 18: 62-63.
- Maalouf NM, Cameron MA, Moe OW, Sakhaee K. Novel insights into the pathogenesis of uric acid nephrolithiasis. Curr Opin Nephrol Hypertens 2004; 13: 181-189.
- Moreira DM, Friedlander JI, Hartman C, Elsamra SE, Smith AD. Using 24-hour urinalysis to predict stone type. J Urol 2013; 190: 2106-2111.
- Chou YH, Li WM, Li CC, Huang SP, Liu CC. Clinical study of uric acid urolithiasis. Kaohsiung J Med Sci 2007; 23: 298-301.
- Sakhaee K. Epidemiology and clinical pathophysiology of uric acid kidney stones. J Nephrol 2014; 27: 241-245.
- Wong YV, Cook P, Somani BK. The association of metabolic syndrome and urolithiasis. Int J Endocrinol 2015; 2015: 570674.
- Li H, Klett DE, Littleton R, Elder JS, Sammon JD. Role of insulin resistance in uric acid nephrolithiasis. World J Nephrol 2014; 3: 237-242.
- Enomoto A, Kimura H, Chairoungdua A. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature 2002; 417: 447- 452.
- Tasic V, Hynes AM, Kitamura K, Cheong HI, Lozanovski VJ. Clinical and functional characterization of URAT1 variants. PLoS One 2011; 6: 28641.
- Stiburkova B, Sebesta I, Ichida K. Novel allelic variants and evidence for a prevalent mutation in URAT1 causing renal hypouricemia: biochemical, genetics and functional analysis. Eur J Hum Genet 2013; 21: 1067-1073.
- Negri AL, Spivacow R, Del Valle E. Clinical and biochemical profile of patients with pure uric acid nephrolithiasis compared with pure calcium oxalate stone formers. Urol Res 2007; 35: 247-251.
- Doshi M, Takiue Y, Saito H. The increased protein level of URAT1 was observed in obesity/metabolic syndrome model mice. Nucleosides Nucleotides Nucleic Acids 2011; 30: 1290-1294.