Research Article - Biomedical Research (2017) Volume 28, Issue 2
Genetic polymorphism of candidate genes for fecundity traits in Egyptian sheep breeds
Ibrahim AH Barakat1,2,*, Lamiaa M Salem1, Niveen M Daoud3,4, Wagdy KB Khalil1, Karima F Mahrous11Department of Zoology, College of Science, King Saud UniversityP.O. Box 2455, Riyadh 11451, Riyadh, Saudi Arabia
2Department of Cell Biology, National Research Center, 33 Bohouth st., Dokki, Giza, Egypt
3Department of Animal Reproduction and Artificial Insemination, National Research Center, 33 Bohouth st, Dokki, Giza, Egypt
4Department of Microbiology, Faculty of Clinical Pharmacy, Taif University, Kingdom of Saudi Arabia
- *Corresponding Author:
- Ibrahim AH Barakat
Department of Zoology, College of Science
King Saud University, Saudi Arabia
Accepted on July 02, 2016
Abstract
In reproductive planning, intervals between lambings, season, age of ewe, heat stress, nutrition state or breed are some of the factors which have a great effect on sheep fertility. Different mutations in the growth differentiation factor 9 (GDF9) and the bone morphogenetic protein 15 (BMP15) genes cause an increase in the ovulation rate in sheep. The aim of the current study was to determine the mutations in GDF9 and BMP15 genes and the probability of their effect on fertility in the major Egyptian sheep breeds. Blood samples were collected using EDTA from the Barki, Ossimi and Rahmani sheep breeds at different locations in Egypt, and DNA was then isolated using the salting out method. The PCR products of the genes were digested with MseI and DdeI restriction enzymes for the GDF9 gene, and with HinfI and DdeI for the BMP15 gene. The results showed that the PCR products digested with restriction enzymes exhibited a substitution in the G7 locus of the GDF9 gene in the Ossimi and Rahmani sheep breeds, and a substitution in the FecGH locus of the GDF9 gene in all sheep breeds. On the other hand, while the BMP15 gene loci digested only with HinfI showed mutations, this did not occur in all samples of the three Egyptian breeds. In conclusion, the BMP15 gene loci showed no polymorphism, while only the GDF9 gene loci were polymorphic in Barki, Ossimi and Rahmani sheep breeds.
Keywords
PCR- RFLP, Polymorphism, GDF9, BMP15, Sheep.
Introduction
There are more than 900 different strains from sheep (Ovise aries) that differ greatly in the fertility and physiological traits, including ovulation rate [1]. In some cases, the difference in ovulation rate may be attributed to the influence of case setting genes [2]. Actually, mutations having great effects on ovulation rate were detected in the genes expressing different proteins, including the growth dierentiation factor 9b (GDF9), BMP receptor-IB (ALK6), transforming growth factor beta (TGFß) superfamily, bone morphogenetic protein (BMP15) and TGFß receptor [3-7]. Moreover, the additional mutations in some of these genes, or in another gene, are similar to be attended in other fertile mammal's strains [2]. Therefore, the useful sheep largely in that it demonstrates that ability to produce two or three offsprings instead of one [8].
Concerning DNA testing for main genes related to fertility and inheritance patterns, scientific research demonstrated that there are main genes have significant possibility to reproductive performance elevation of sheep worldwide [2]. Three genes related to fertility were discovered in sheep, Growth differentiation factor 9 (GDF9) known as FecG, bone morphogenetic protein receptor type 1B (BMPR1B) known as FecB and bone morphogenetic protein 15 (BMP15), known as FecX [4,6]. In the Rasa aragonesa sheep strain, six various mutations were discovered for BMP15 gene; FecXI, FecXH, FecXL, FecXG, FecXB and FecXR [3,4,9,10], and each one of them has a main effect on fertility. The sterile ewes have two inactive copies from BMP15 gene (homozygous animals) [3,4]. While, ewes that have inactive single copy from the same gene (heterozygous animal) were fertile and have higher ovulation rate and lambing [3,4,11,12]. As for the GDF9 gene, it has been identified on eight different mutations (G1-G8). Three of them (G2, G3 and G5) are nucleotide changes which did not lead to change in the amino acids. Other remaining nucleotides (G1, G4, G6, G7, and G8) resulted in changes in amino acids. The first polymorphism (G1), that changes arginine to histidine in exone 1, replaces a basic charged polar group with another group for mature peptide at a position of furin cleavage. So, it does not affect the activity of mature protein [4]. Ewes containing one allele (heterozygote) from two copy of mutated GDF9 (FecGH) gene is fertile and has high rate of ovulation [4]. On the other side, ewes containing the same alleles (homozygote) for this mutation are infertile in addition to the elementary ovary failure. Moreover, the heterozygous ewes for mutation of GDF9 and BMP15 genes are fertile and these mutations have additional effects on ovulation rate [4].
In Egypt, Barki, Ossimi and Rahmani are major sheep breeds. They are characterized by fat tail, coarse wool, and small to medium size. Body size varies between 51-53 kg adult weight for the Ossimi and Rahmani breeds, and 44 kg for the Barki breed. The annual milk yield ranges between 65 kg for the Ossimi and Rahmani breeds and 59 kg for the Barki breed [13]. Until now, no selection programmes have succeeded in increasing the litter size of Egyptian sheep breeds because of the low possibility of genetic transition. The genetic decline for litter size trait indicates that the addition of genetic variation is not the main component of genetic variation phenotypic [13]. Assistance and promotive molecular genetic studies and biological aspects associated used to study and understand the basic genes. There are reports about the effect of genes on the litter size in other strains of sheep, indicating the ability to search for individual mutations that may affect the characteristics of sheep. Therefore, the objective of this study was to detect the presence of polymorphism in the FecXG and FecXB loci for the BMP15 gene, and the FecGH and FecG7 loci for the GDF9 gene in the Barki, Ossimi and Rahmani Egyptian sheep breeds, to determine their role in the enhancement of the fertility rate of these breeds.
Materials and Methods
Animals
Multiparous Rahmani (3-4 years), Barki (3-4 years) and Ossimi (2-3 years) ewes were used for this experiment. The Barki breed was grown at faculty of Agriculture, Cairo University. The Ossimi breed was grown in different places in the Nile Valley and Delta, and Rahmani breed was sourced from Beheira province in the northern part of the Nile Delta.
Chemicals
All chemicals that were used to isolate DNA were purchased from Sigma Chemical Co. (St. Louis, MO, USA) unless otherwise noted. Primers and restriction enzymes were purchased from Fermentas Co., Germany.
Sample collections
One hundred and twenty six unrelated female Egyptian sheep with a range of ages (Barki; n=50, Ossimi; n=36 and Rahmani; n=40) were used to collect the blood samples for DNA isolation.
DNA isolation
The blood samples collected from sheep vein were put in a sterile tube containing 0.5 ml of 0.5 M ethylenediaminetetraacetic acid (EDTA). Salting out method was used to isolate DNA from whole blood [14]. Isolated DNA was kept on at -20°C until used after dissolving in TE buffer. Spectrophotometer and agarose gel electrophoreses were used to evaluate the quality and quantity of isolated DNA. The concentration of the evaluated DNA was adjusted at 100 ng/μl and was used as a template in polymerase chain reaction (PCR).
Polymerase chain reaction (PCR) process
PCR master mix (MM) that consists of 0.2 mM dNTPs (Biotechnology, Cairo, Egypt), 50 mM KCl (Ran Baxy, New Delhi, India), 10 mM Tris (pH 9), 1.5 mM MgCl2 (Sigma), 0.1% Triton X-100 (Merk), 1.25 units of Taq polymerase (Bioron, Germany), 0.01% gelatin (Merk) and 1.0 μM forward and reverse of specific primers (Table 1). The MM was divided into PCR tubes each containing 100 ng of DNA. The PCR cycle was adjusted at 94°C for 5 min, followed by 35 cycles of 94°C for 45 s, 58°C for 40 s, 72°C for 1 min and a final extension at 72°C for 10 min. The PCR product was subjected to electrophoresis for detection amplified DNA.
Restriction fragment length polymorphism analysis (RFLP) technique
The PCR products were digested with restriction enzymes specific for each gene (Table 1) at 25 μl final reaction volume. The RFLEP was carried out using 20 μl from digested PCR product and was incubated overnight at 37°C. After digestion time, the restricted product was analysis by electrophoresis and photographed using gel documentation system. Labimag software downloaded from the company web page: www.labimage.com was used to detect the fragment sizes and alleles.
| Gene | Site | Size (bp) | Sequences (5'-3') | R. E | References |
|---|---|---|---|---|---|
| BMP15 | FecXG | 141 | CACTGTCTTCTTGTTACTGTATTTCAATGAGAC GATGCAATACTGCCTGCTTG |
Hinf1 | [4] |
| FecXB | 153 | GCCTTCCTGTGTCCCTTATAAGTATGTTCCCCTTA TTCTTGGGAAACCTGAGCTAGC |
DdeI. | [4] | |
| GDF9 | FecGH | 139 | CTTTAGTCAGCTGAAGTGGGACAAC ATGGATGATGTTCTGCACCATGGTGTGAACCTGA |
DdeI | [4] |
| G7 | 158 | CAGTATCGAGGGTTGTATTTGTGTGGGGCCT GCCTCTGGTTCCAGCTTCAGTC |
MseI. | [4] |
Table 1. Primer sequence of candidate genes for prolificacy in Egyptian sheep breeds.
Results
DNA fragments of the GDF9 and BMP15 genes were digested using four different restriction enzymes (MseI and DdeI for GDF9 and HinfI and DdeI for BMP15 loci genes) in order to detect the genetic variability between the three Egyptian sheep breeds. One hundred thirty nine and one hundred fifty eight base pair DNA fragments from exon 1 of the GDF9 gene, also 141 and 153-bp from exon 2 of the BMP15 gene were successfully amplified. Genotypes of each individual sheep breed were detected using horizontal electrophoresis (Figures 1-4).
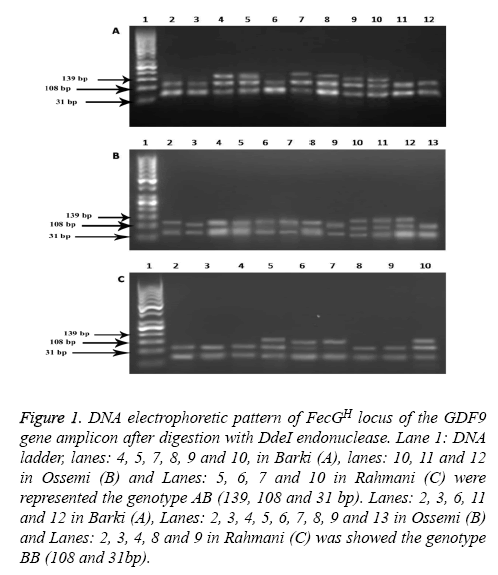
Figure 1 : DNA electrophoretic pattern of FecGH locus of the GDF9 gene amplicon after digestion with DdeI endonuclease. Lane 1: DNA ladder, lanes: 4, 5, 7, 8, 9 and 10, in Barki (A), lanes: 10, 11 and 12 in Ossemi (B) and Lanes: 5, 6, 7 and 10 in Rahmani (C) were represented the genotype AB (139, 108 and 31 bp). Lanes: 2, 3, 6, 11 and 12 in Barki (A), Lanes: 2, 3, 4, 5, 6, 7, 8, 9 and 13 in Ossemi (B) and Lanes: 2, 3, 4, 8 and 9 in Rahmani (C) was showed the genotype BB (108 and 31bp).
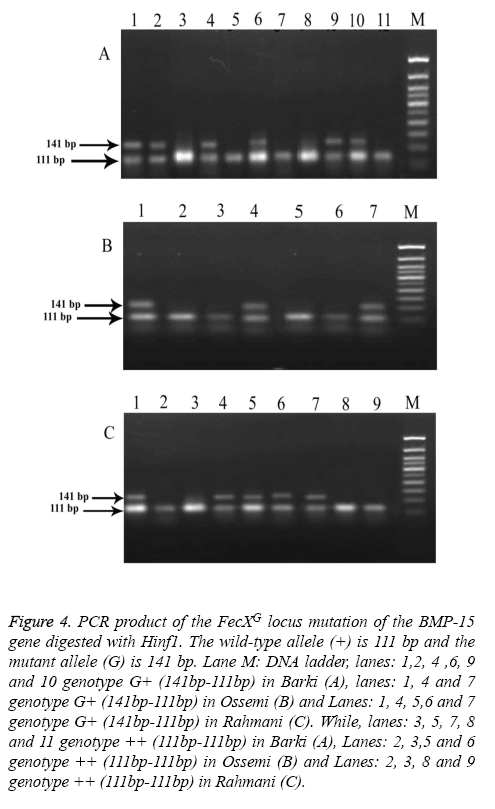
Figure 4: PCR product of the FecXG locus mutation of the BMP-15 gene digested with Hinf1. The wild-type allele (+) is 111 bp and the mutant allele (G) is 141 bp. Lane M: DNA ladder, lanes: 1,2, 4 ,6, 9 and 10 genotype G+ (141bp-111bp) in Barki (A), lanes: 1, 4 and 7 genotype G+ (141bp-111bp) in Ossemi (B) and Lanes: 1, 4, 5,6 and 7 genotype G+ (141bp-111bp) in Rahmani (C). While, lanes: 3, 5, 7, 8 and 11 genotype ++ (111bp-111bp) in Barki (A), Lanes: 2, 3,5 and 6 genotype ++ (111bp-111bp) in Ossemi (B) and Lanes: 2, 3, 8 and 9 genotype ++ (111bp-111bp) in Rahmani (C).
Genetic polymorphism of the GDF9 gene
The PCR products of the GDF9 (FecGH) gene digested with DdeI are shown in Figure 1. The results for genotype AB found that all sheep breeds showed mutations in which the restriction enzyme DdeI digested the sequences of GDF9 (FecGH) to three fragments (139, 108 and 31bp). For genotype BB, however, the restriction enzyme DdeI digested the sequences of GDF9 (FecGH) to only two fragments (108 and 31bp) in Barki, Ossemi and Rahmani breeds. Through different Egyptian strains (Table 2); the present results showed that strains just contained two genotypes (AB and BB), with the Barki breed having a high frequency of the BB genotype, followed by Rahmani and Ossimi. While, the Barki breed had lower frequency of A allele and AB genotype when compared with the Ossimi and Rahmani breeds.
| Breed | No. of Animals | Allele frequency | Genotype frequencies | |||
|---|---|---|---|---|---|---|
| A | B | AA | AB | BB | ||
| Barki | 50 | 0.11 | 0.89 | 0 | 0.22 | 0.78 |
| Ossimi | 36 | 0.23 | 0.77 | 0 | 0.46 | 0.54 |
| Rahmani | 40 | 0.18 | 0.82 | 0 | 0. 36 | 0.64 |
Table 2. Allele and genotypes frequencies of the GDF9 (FecGH) gene among tested Egyptian sheep breeds.
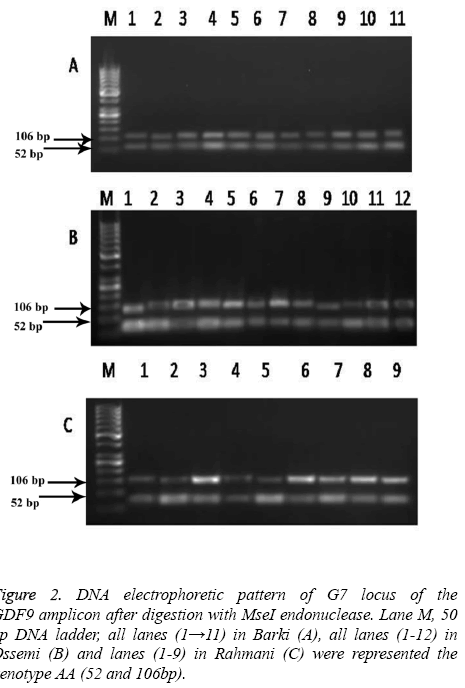
The high frequency of allele B and A was found in Barki and Ossimi breeds, respectively. The PCR products of the GDF9 (FecG7) gene digested with MseI showed a mutation which enabled the restriction enzyme to digest the gene (Figure 2). The Barki breed alleles of this gene, however, had no restriction site and lead to one fragment 158 bp of DNA (Figure 2A). The Ossimi breed allele types of the same gene, meanwhile, had one restriction site and resulted in two DNA fragments of 52, 106 bp (Figure 2B). The same trend was observed in the Rahmani sheep breed, in that the alleles of this gene had one restriction site and lead to two DNA fragments of 52 and 106 bp (Figure 2C).
Genetic polymorphism of the BMP15 gene
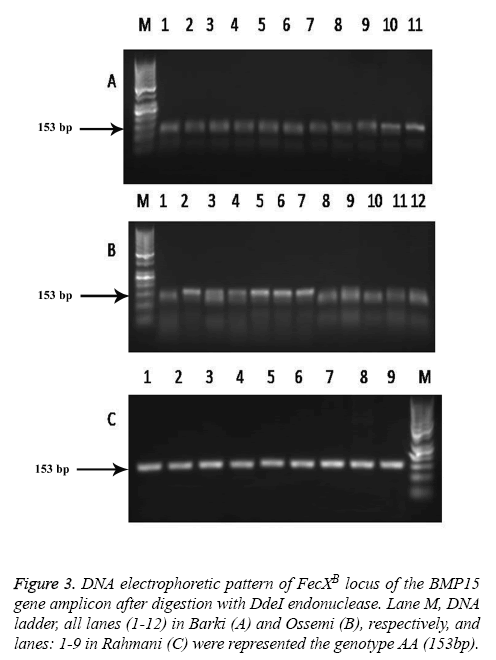
The results revealed that there was no polymorphism of the BMP15 (FecXB) gene in any of the surveyed breeds of Egyptian sheep (Figure 3), the same previous results were found in all digested PCR products with the DdeI restriction enzyme (one band, 153 bp). The digested gene products of PCR BMP15 (FecXG) with the HinfI restriction enzyme did show mutation, however, but not in all samples (Table 3 and Figure 4). Most of the PCR products of Barki, Ossimi and Rahmani breeds digested with HinfI restriction enzyme resulted in 141bp and 111bp fragments (Figures 4A-4C, genotype G+), but, in the majority of the samples from the Ossimi breed, genotype ++ of the BMP15 (FecXG) gene showed as only one band in the agarose gel electrophoresis (Figures 4A and 4C). This outcome was much less common, however, with the Barki and Rahmani breeds. Of the different tested breeds (Table 3), the results revealed that only two genotypes (++ and G+) were found in all breeds. A high frequency of genotype ++ was found in the Barki breed, while a frequency of G+ was found in the Rahmani breeds. There was not much variation between the breeds, however, in the frequency of the ++ allele. Moreover, the frequency of the + and G allele was high in the Barki and Rahmani sheep breeds, respectively, although the Ossimi breed had middle values for allele and genotype frequency.
| Breed | No. of animals | Allele frequency | Genotype frequencies | |||
|---|---|---|---|---|---|---|
| G | + | GG | G+ | ++ | ||
| Barki | 50 | 0.34 | 0.66 | 0 | 0.68 | 0.32 |
| Ossemi | 36 | 0.36 | 0.64 | 0 | 0.72 | 0.28 |
| Rahmani | 40 | 0.38 | 0.62 | 0 | 0.76 | 0.24 |
Table 3. Allele and genotypes frequencies of the BMP15 (FecXG) gene between the different Egyptian sheep breeds.
Discussion
Genotyping was carried out using PCR-RFLP, because it's a rapid, exact and simple technique, to prolife sheep and goats [15-17]. In the present study, this technique was used to detect the genetic polymorphism according to Hanrahan et al. [4]. Polymorphism results concerning the GDF9 (FecG7 and FecGH) gene confirmed previous observations that had been reported by Hanrahan et al. [4]. The present study in Barki, Ossimi and Rahmani sheep showed same results to those obtained previously [4,18-22]. Most of the tested animals in the present study were of a heterozygous genotype and had a high fertility rate, which is in agreement with Kasiriyan et al. [23], who reported the increased fecundity rate of the GDF9 gene mutant in Sangsari sheep strain. GDF9 mutations increased the ovulation rates in heterozygous ewes, while the homozygous ewes were sterile as a result of failure of follicular development [3,4,11]. Polley et al. [24] have been identified 8 different mutations in exons 1 and 2 of GDF9 gene. Mutation named Thoka gene in GDF9 gene was found in Icelandic sheep [25]. Another mutation detected by Silva et al. [26] in the Brazilian Santa Inês sheep was called Embrapa. There is a mutation in allele A (G to A), called G1 which was detected in Iranian sheep breeds such as Mehraban breed [27], Kordi and Arabi breeds [28], and Baluchi breed [29]. No homozygous genotype (AA) was found [30], because this genotype may be lethal or has sterility effects, while was found in other breeds such as Mehraban and Baluchi sheep breeds as reported by Abdoli et al. [27] and Moradband et al. [29], repectively. On the other hand, Yadollah et al. [31] showed that there is no any mutation and polymorphism in the GDF9 gene loci. In general, Ala and Rafat [32] reported that many different loci are under investigation between sheep breeds, more than genetic background. Our results were similar to those of Borhan [33] who found that is no genetic polymorphism in BMP15 gene in 200 goat breeds. Also, Hua et al. [34] showed that there is a 99% similarity with goat for BMP15 gene. Moreover, no mutation detected in Chios, Awassi, Kivircik and Imrose sheep breeds for FecXH, FecXb and FecXI gene loci [35]. On the other hand, different goat breeds showed two polymorphic genotypes in exon 2 of BMP15 gene [36-39]. The detected mutations for BMP15 (FecXG) and BMP15 (FecXB) in our study had no effect on litter size (genotype GG) in Barki, Ossimi and Rahmani sheep breeds. The same previous results were reported by Moradband et al. [40] in Baluchi Sheep, Davis et al. [21] in East Friesian, Blueface Leicester, Romanov, D’Man, German White-headed Mutton, Loa, Barbados Blackbelly, Teeswater, Finn, Chios, Mountain, Lleyn and Galician sheep breeds, Kumar et al. [15] in Mulpura sheep and Guan et al. [16] in Charolais, Romney Hills, Suffolk, Chinese Merino and Dorset sheep breeds. In all these breeds the allele of FecB mutatnt was not segregated. In addition, the genetic polymorphism analysis showed that there is no relationship between twining rates and mutated alleles at the BMP15 FecXG and BMP15 FecXB genes loci in Baluchi sheep breed [24]. In agreement with these findings, the fertility rate in Ossimi, Rahmani and Barki sheep breeds are not related to the mutations of the BMP15 (FecXG) and BMP15( FecXB) loci, which were absent in their genome. It should be attempted to study the other SNPs for these genes loci which may be responsible for the fertility in Egyptian sheep breeds.
Conclusion
The BMP15 gene loci showed no polymorphism, while only the GDF9 gene loci were polymorphic in Barki, Ossimi and Rahmani sheep breeds. These findings were accompanied with the fertility rate of the Egyptian breeds, where the fertility rate in Ossimi, Rahmani and Barki sheep breeds are related to the GDF9 (FecG7 and FecGH) gene but not related to the mutants of the BMP15 (FecXG) and BMP15 (FecXB) loci, which were absent in the genome of these breeds.
Acknowledgement
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at king Saud University for its funding this Research group NO (RG -1435-058).
References
- Terril CF. The distribution of breeds as related to domestication and development of modern genotypes, in: Foote W.C., Bunch D.T. (Eds.), The domestication of sheep; their ancestors, geography, time period and people involved, Proceedings of a Workshop by The International Sheep and Goat Institute, Utah State University, Logan; 1979.
- Davis GH. Major genes affecting ovulation rate in sheep. Genet Sel Evol Suppl 2005; 137: S11-S23.
- Galloway SM, McNatty KP, Cambridge LM, Laitinen MPE, Juengel JL, Jokiranta S, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, Beattie AE, Davis GH, Ritvos O. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet 2000; 25: 279-283.
- Hanrahan JP, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, Galloway S. Mutations in the genes for oocyte derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol Reprod 2004; 70: 900-909.
- Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, Pisselet C, Riquet J, Monniaux D, Callebaut I, Cribiu E, Thimonier J, Teyssier J, Bodin L, Cognie Y, Chitour N, Elsen JM. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation in Booroola ewes. Proc Natl Acad Sci USA 2001; 98: 5104-5109.
- Souza CJ, MacDougall C, Campbell BK, McNeilly AS, Baird DT. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. J Endocrinol 2001; 169: R1-R6.
- Wilson T, Wu XY, Juengel JL, Ross IK, Lumsden JM, Lord EA, Dodds KG, Walling GA, McEwan JC, O’Connell AR, McNatty KP, Montgomery GW. Highly proli?c Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol Reprod 2001; 64: 1225-1235.
- McNatty KP, Juengel JL, Wilson T, Galloway SM, Davis GH, Hudson NL, Moeller CL, Cran?eld M, Reader KL, Laitinen MPE, Groome NP, Sawyer HR, Ritvos O. Oocyte derived growth factors and ovulation rate in sheep. Reprod 2003; 61: 339-351.
- Bodin L, Lecerf F, Pisselet C, San Cristhal M, Bibe M, Mulsant P. How many mutations are associated with increased ovulation rate and litter size in progeny of acaune meat sheep? In Proceeding of International Workshop on Major Genes and QTL in Sheep and Goats. INRA, Toulouse, France; 200, 32-11.
- Monteagudo LV, Ponz R, Tejedor MT, Lavina A, Sierra I. A 17 bp deletion in the bone morphogenetic protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed. Anim Reprod Sci 2009; 110: 139-146.
- Davis GH, McEwan JC, Fennessy PF, Dodds KG, Farquhar PA. Evidence for the presence of a major gene influencing ovulation rate on the X chromosome of sheep. Biol Reprod 1991; 44: 620-624.
- Davis GH, Dodds KG, McEwan JC, Fennessy PF. Live weight, fleece weight and prolificacy of Romney ewes carrying the Inverdale prolificacy gene (FecXI) located on the X-chromosome. Livest Prod Sci 1993; 34: 83-91.
- Almahdy H, Tess MW, El-Tawil E, Shehata E, Mansour H. Evaluation of Egyptian sheep production systems: II. Breeding objectives for purebred and composite breeds. J Anim Sci 2000; 78: 288-295.
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
- Kumar S, Kolte AP, Mishra AK, Arora AL, Singh VK. Identification of the FecB mutation in Garole × Malpura sheep and its effect on litter size. Small Rumin Res 2006; 64: 305-310.
- Guan F, Shou-ren L, Shi GQ, Yang LG. Polymorphism of FecB gene in nine sheep breeds or strains and its effects on litter size, lamb growth and development. Anim Reprod Sci 2007; 99: 44-52.
- Polley S, De S, Brahma B, Mukherjee V, Batabyal S, Arora JS, Pan S, Samanta AK, Datta TK, Goswami SL. Polymorphism of BMPR1B, BMP15 and GDF9 fecundity genes in prolific Garole sheep. Trop Anim Health Prod 2009; 42: 985-993.
- Juengel JL, Hudson NL, Whiting L, Natty KPM. Effects of immunization against bone morphogenetic protein 15 and growth differentiation factor 9 on pregnancy in Ewes. Biol Reprod 2004; 70: 557-561.
- Chu MX, Wang S, Fang L. Association between PCR-SSCP of growth differentiation factor 9 gene and high prolificacy in small tail han sheep. Anim Biotechnol 2004; 15: 111-120.
- Chu MX, Liu ZH, Giao CL, He YQ, Fang L, Ye SC, Chen GH, Wang GY. Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries). J Anim Sci 2007; 85: 598-603.
- Davis GH, Balakrishnan L, Ross IK, Wilson T, Galloway M, Lumsden BM, Hanrahan JP, Mullen MX, Mao Z, Wang GL, Zhao ZS, Robinson JJ, Mavrogenis AP, Papachristoforou C, Peter C, Baumung R, Cardyn P, Boujenane I, Cockett NE, Eythorsdottir E, Arranz JJ, Notter DR. Investigation of the Booroola (FecB) and Inverdale (FecXI) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim Reprod Sci 2006; 92: 87-96.
- Liao WX, Moore RK, Shimasaki S. Functional and molecular characterization of naturally occurring mutations in the oocyte secreted factors bone morphogenetic protein-15 and growth and differentiation factor-9. J Boil Chem 2004; 17: 17391-17396.
- Kasiriyan MM, Hafezian SH, Hassani N. Genetic polymorphism BMP15 and GDF9 genes in Sangsari sheep of Iran. International Journal of Genetics and Mol Biol 2011; 3: 31-34.
- Polley S, De S, Brahma B, Mukherjee A, Vinesh PV, Batabyal S, AroraJ S, Pan S, Samanta AK, Datta T K, Goswami SL. Polymorphism of BMPR1B, BMP15 and GDF9 fecundity genes in prolific Garole sheep. Trop Anim Health Prod 2010; 85: 122-129.
- Nicol L, Bishop SC, Pong-Wong R, Bendixen C, Holm LE, Rhind SM, Mc-Neilly SA. Homozygosity for a single base-pair mutation in the oocyte specific GDF9 gene results in sterility in Thoka sheep. Reproduction 2009; 138: 921-933.
- Silva BD, Castro EA, Souza CJ, Paiva SR, Sartori R, Franco MM, Azeved HC, Silva TASN, Vieira AMC, Neves JP, Melo EO. A new polymorphism in the Growth and Differentiation Factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep. Anim Genet 2011; 42: 89-92.
- Abdoli R, Zamani P, Deljou A, Rezvan H. Association of BMPR-1B and GDF9 genes polymorphisms and secondary protein structure changes with reproduction traits in Mehraban ewes. Gene 2013; 524: 296-303.
- Ghaderi A, Beigi NMT, Mirzadeh KH, Fayazi J, Sadr AS. Identification of the GDF9 mutation in two sheep breeds by using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique. Afric J Biotech 2010; 9: 8020-8022.
- Moradband F, Rahimi G, Gholizadeh M. Association of polymorphism in fecundity genes of GDF9, BMP15 and BMPR-1B with litter size in Iranian Baluchi sheep. Asian-Aust J Anim Sci 2011; 9: 1179-1183.
- Pouya Z, Ramin A, Ali D, Hosein R. Polymorphism and bioinformatics analysis of growth differentiation factor 9 gene in Lori sheep. Ann Anim Sci 2015; 2: 337-348.
- Yadollah B, Sajad B, Hamid RM, Vaheid CA, Seyed AM. The Polymorphism of GDF-9 Gene in Hisari Sheep. Biol Forum Int J 2014; 6: 46-52.
- Ala NF, Rafat A. Genetic Polymorphism of GDF9 Gene in Iranian Moghani Sheep Breed. Iranian J Appl Animal Sci 2014; 4: 887-890.
- Borhan S. Investigation of BMP15 gene polymorphisms associated with twining in Markhoz goat. Biharean Biologist 2015; 9: 1-4.
- Hua GH, Chen SL, Ai JT, Yang LG. None of polymorphism of ovine fecundity major genes FecB and FecX was tested in goat. Animal Reproduct Sci 2008; 108: 279-286.
- Gursel FE, Akis A, Durak H, Mengi A, Oztabak K. Determination of BMP-15, BMPR-1B and GDF-9 Gene Mutations of the Indigenous Sheep Breeds in Turkey. Kafkas Kafkas Üniversitesi Veteriner Fakültesi Dergisi 2011; 17: 725-729.
- Cailan, J, Ming-Xing C, Jinyu W, Li F, Sucheng Y. Cloning and sequence analysis of exon 2 of BMP15 gene in Jining Grey Goats. Chinese J Animal Vet Sci 2007; 34: 52-55.
- Chu MX, Jiao CL, He YQ, Wang JY, Liu ZH, Chen GH. Association between PCR-SSCP of bone morphogenetic protein 15 gene and prolificacy in Jining Grey goats. Animal Biotechnol 2007; 18: 263-274.
- Wang Y, Yuanxiao L, Zhang N, Wang Zh, Bai J. Polymorphism of Exon 2 of BMP15 Gene and Its Relationship with Litter Size of Two Chinese Goats. Asian-Australian J Animal Sci 2011; 24: 905-911.
- Abdel-Rahman SM, Mustafa YA, Abd Errasool HA, El Hanafy AA, Elmaghraby AM. Polymorphism in BMP-15 gene and its association with litter size in Anglo-nubian goat. Biotechnol Animal Husbandry 2013; 29: 675-683.
- Moradband F, Rahimi G, Gholi M. Association of Polymorphisms in Fecundity Genes of GDF9, BMP15 and BMP15-1B with Litter Size in Iranian Baluchi Sheep. Asian-Australasian J Anim Sci 2011; 24: 1179 -1183.