Research Article - Biomedical Research (2019) Volume 30, Issue 2
Evaluation of clonal relatedness among different sources of E. coli isolates in Iranian children with urinary tract infection (UTI) and age-matched healthy people.
Shahin Najar Peerayeh1, Masoumeh Navidinia2*, Fatemeh Fallah3, Bita Bakhshi1 and Masoud Alebouyeh4
1Department of Bacteriology, Tarbiat Modares University, Tehran, IR Iran
2School of Allied Medical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, IR Iran
3Pediatric Infection Research Center, Research Institute For children Health, Shahid Beheshti University of Medical Sciences, Tehran, IR Iran
4Research Center of Gastroenterology and Liver Diseases Shahid Beheshti, University of Medical Sciences, Tehran, IR Iran
- *Corresponding Author:
- Masoumeh Navidinia
School of Allied Medical Sciences
Shahid Beheshti University of Medical Sciences
Tehran, IR Iran
Accepted date: February 4, 2019
DOI: 10.35841/biomedicalresearch.30-19-035
Visit for more related articles at Biomedical ResearchAbstract
Urinary tract infections (UTIs) currently rank among the most prevalent infectious diseases worldwide, with chronic and recurrent infections being especially problematic. Uropathogenic E. coli (UPEC), which is a causative agent in most cases of UTIs, expresses a multitude of virulence factors. Some virulence factors and specific genes were examined by PCR method. Genetic diversity was evaluated by phylogenetic typing groups. To know the genetic linkages among various E. coli, we evaluated clonal relatedness among different sources of E. coli isolates in Iranian children with UTI and age-matched healthy people by PFGE. Some pathogenicity determinants were more prevalent in urinary strains rather than fecal E. coli strains, significantly. There were PAI IICFT073, PAI II J96, PAI I536, PAI ICFT073, PAIII536, PAI IV, gafD, focG, vat, usp, hlyD, sat, cnf1, picU, fliC (H7), kpsMTII, kpsMTIII. UPEC were mainly found in phylogenetic typing groups B2 and D, while in fecal isolates, phylogenetic groups A and D were the most common. One hundred and fifty E. coli genomic DNA were digested by XbaI restriction enzyme and subjected to PFGE. A high level genetic diversity of DNA restriction pattern was reported. So, different clonalities were observed among different sources of E. coli isolates in Iranian children with UTI and age-matched healthy people. With attention to these results, genetic patterns showed that the strains had different clonalities. The differences in the PFGE pattern are a result of various genetic events caused by consistent changes with some independent genetic events. Lack of genetic similarity among E. coli isolates with different sources can be explained by the acquisition of different gene factors.
Keywords
Clonal relatedness, Fecal E. coli isolates, Healthy people, Iranian children, Urinary tract infection (UTI), Uropathogenic E. coli (UPEC).
Introduction
UTI with chronic and recurrent infection is common in childhood around the world. It can have wide range complications, including kidney damage, kidney failure, or blood poisoning (sepsis) [1,2]. UTI occurs in about 3-5% of girls and 1% of boys less than 5 years. After the first incidence of UTI, 60%-80% girls get a second infection within 18 months old [3]. In adults, there are some evidences that about 40% women (in their early 20s) and 12% men (above 85) will experience UTI symptomatically with increasing incidences. During 6 to 12 months, about 25% of these women will get infected again. Uropathogenic E. coli (UPEC) is the primary etiologic agent associated with UTIs [2]. UPEC can invade various types of bladder cells, such as the stratified layers of the bladder urothelium. Both the establishment and persistence of UPEC in the urinary tract is facilitated by invasion of the host cell [4]. UPEC persists, with interior urothelial cells lying in a dormant state within late endosomal vesicles. In addition, UPEC isolates chiefly carry individual virulence factors that help to colonize successfully in the mammalian urinary tract and can cause intestinal and extra-intestinal infection [2,4,5]. Furthermore, UPEC isolates possess several virulence factors which are uncommon in fecal E. coli isolates. The mechanisms underlying the evolution and emergence of such new bacterial pathogens are not well understood, and these atypical strains now are attracting more attention. The variety of virulence determinants due to horizontal gene transfers of mobile elements, such as pathogenicity islands (PAIs), phages and plasmids contributing to a strain’s pathogenic potential [6-10].
By applying PFGE, discretion of alteration in great pieces of DNA is possible. It is also a very effective tool in epidemiological studies and gene mapping of microbes. In a monotonous magnetic field, segments more than 50 kb length transmit a direction across the gel and move in a zigzag pattern. DNAs separations as bands carry out better across the gel. In PFGE method, closely related bacterial species display patterns equivalent to DNA separation, and those which do not possess similarity or are less similar manifest various separation patterns. Therefore, these attributes can be used to identify the common bacterial agent of a disease. PFGE can also be employed to monitor and evaluate various microorganisms in clinical specimens and existent ones in soil and water. In the recent years, PFGE has proven to be an extremely remarkable and powerful tool for control, prevention, and surveillance of infections in several populations [11]. At first, PFGE illustrated by Schwartz in 1983 with the intermittent change of electricity current brings DNA migration. Recently, several techniques have been expanded to recognize and classify bacteria: however, all these methods are not appropriate. Otherwise, PFGE is the applicable method for detection, identification, and classification of genome of microorganisms accurately and sensitively. Kindergartens, schools, hospitals, prisons, and even homeless refuges are exposed to a greater danger of infection outbreak due to bacteria. Moreover, in some places, such as hospitals, there are patients with weakened immune systems: diseases are can spread quickly there. Hospitals are places where the possibility of introducing bacteria into surgery wounds, in vitro fertilization (IVF), and catheterization is possible. Determination of the exact time in suspicious cases of an outbreak is important because the outbreak source can be a hospital worker or hospital food. PFGE method gives researchers the ability of linear DNAs separation longer than 10 mega base pair (Mbp) [12]. Also, PFGE is a useful instrument to determine the relationship between several strains of a single species; to approximate the size and chromosomal mechanism; to illustrate and clarify the genome of eukaryotes and prokaryotes; to assist physical gene mapping; to permit large DNA fragment analysis; and to allow for primary origin detection of genes in fungi, protozoa, bacteria, and even mammals [13]. Albeit, PFGE discriminatory power has evaluated approval in compare with other subtyping methods [14-16]. So, the aim of this study is the evaluation of clonal relatedness among different sources of E. coli isolates in Iranian children with UTI and age-matched healthy people by PFGE method.
Materials and Methods
Bacterial isolates
This was a retrospective study wherein we considered 150 E. coli strains. We selected two groups. First of all, 100 E. coli strains were isolated from urine (50 cases) and stool (50 cases) samples collected from 2-12 year old children with UTI: these were all patients referred to the nephrology ward of the Mofid Children Hospital. From all children with UTI, we isolated E. coli from their stool specimen. We also isolated 50 E. coli from stool samples of age-matched healthy people. Urine samples were collected in the sterile containers and were cultured onto MacConkey agar, sheep blood agar, and Muller-Hinton agar for colony counting. After incubating at 37°C overnight, we examined microorganisms’ growth. Each plate with growth more than 105 CFU/ml was processed subsequently. Besides, 100 E. coli which were isolated from stool samples of UTI patients (50 strains) and healthy children as a control group (50 strains) were cultured on MacConkey agar plates and incubated at 37 °C overnight. In total, all E. coli strains were approved biochemically and stored in Luria-Bertani (LB) broth supplemented with sterilized 15% glycerol at -20°C for further examination (all media was purchased from Sigma Aldrich Company, Germany) [7]. Some virulence factors and specific genes were examined by PCR method. Genetic diversity was evaluated by phylogenetic typing groups [17].
PFGE
We used a standard operating protocol for PFGE [16]. Plugs were made ready from bacterial suspensions accompanied by proteinase K (10 μl at 20 mg/ml) treatment. The plugs were lysed in ES buffer, including 0.5 M EDTA, pH 9.0, and 1% sodium-lauryl-sarcosine. After treatment with proteinase K at 54°C for 1 to 2 hours, plugs were washed two times (for 15 min each time at 50°C) in sterile distilled water and four times (for 15 minutes each time) in TE buffer (Triss-EDTA). Slices of plug with 2 mm thickness were digested with XbaI endonuclease. CHEF-DRII PFGE apparatus (Bio-Rad) was performed for PFGE by separation on a 1% agarose gel (Seakem) into 0.5% tris-borate-EDTA buffer at 14°C and 6 V/cm. PFGE was done by Bio-Rad CHEF-DRIII system and condition was reported as: digestion temp (°C): 37; digestion time (h): 4-16; units of enzyme per plug: 30; run time (h): 20; initial switch time (s): 2.2; and final switch time (s): 54.2.+
Salmonella cholerasuis serotype Branderup H9812 was used as the molecular size marker. The gels were stained with ethidium bromide and photographed under ultraviolet light. Calculation of phylogeny and drawing of phylogenetic tree (phenograms) unweighted paired group (UPGMA) method by BioNumerics Gelcompar II software version 4.0 (Applied Maths, Sint- Martens-Latem, Belgium) and band-based Dice Similarity Coefficient (DSC) was used Interpretation was done by using the guidelines set out previously [16-19].
Data analysis
Continuous variables were expressed as mean ± Standard Deviation (SD). Discrete variables were reported as frequency and percentage. Chi square test and Fisher's exact test were used to access the relation between quantities' variables. For all statistical analyses, a p-value of <0.05 was considered to be significant. Statistical analysis was conducted using the SPSS version 24. χ2 test was used for ratio comparison of the same groups (fecal isolates derived from healthy people and UTI patients). McNamara test was used for ratio comparison of the different groups (fecal isolates and urine isolates derived from UTI patients).
Results
Of the 50 fecal E. coli isolates, 44% fell into group A, 16% into B2, 14% in B1, and 32% into D, and urinary isolates 34% fell into group D, 54% into B2, 4% in B1, and 8% into A. Of the 50 fecal E. coli isolates (control), 48% fell into group A, 6% into B2, 26% in B1, and 22% into D. According to data presented in Table 1, the distribution of fecal E. coli isolates and urinary isolates among the four phylogenetic groups is as following: Distribution PAI genes, such as PAI ICFT073 (74 vs. 26%), PAI IICFT073 (38 vs. 14%), PAI I536 (36 vs. 6%), PAI IV536 (86 vs. 42%), PAI II J96 (30 vs. 10%) were more frequent virulence markers in urinary isolates than fecal E. coli and PAIII536 (22 vs. 4%), PAI III536 (6 vs. 0%), PAI I J96 (4 vs. 0%) markers was almost similar in urinary isolates and fecal E. coli. Distribution of adhesion genes, such as bmaE (16 vs. 6%), gafD (20 vs. 2%), focG (22 vs. 6%) were more frequent virulence markers in urinary than fecal E. coli and afa (6 vs. 10%), fimH (92 vs. 98%) markers were almost similar in both isolates. Distribution of toxin genes, such as vat (96 vs. 4%), usp (54 vs. 6%), hlyD (26 vs. 2%), cdtB (18 vs. 10%), sat (44 vs. 8%), cnf1 (26 vs. 0%), picU (42 vs. 2%) were more frequent virulence markers in urinary isolates than fecal E. coli. cvaC (20 vs. 66%) was most frequent marker in fecal E. coli. Distribution of miscellaneous genes, such as fliC (H7 (26 vs. 10%)), ompT (62 vs. 58%) were more frequent virulence factors in urinary than fecal E. coli, and ibeA (14 vs. 26%) was almost similar in both of them. Distribution of protectins genes, such as kpsMTII (70 vs. 46%) was more frequent virulence gene in urinary rather than fecal E. coli. The prevalence of kpsMTI (K1) (46 vs. 54%), kpsMTIII (14 vs. 2%), rfc (O4 LPS) (6 vs. 2%), were almost similar in both of them.
| Genes | Fecal E. coli control N=50 | Fecal E. coli patients N=50 | Urinary E. coli N=50 | p-value |
|---|---|---|---|---|
| PAI genes | ||||
| PAI III 536 | 1 (2%) | 0 (0%) | 3 (6%) | 0.324 |
| PAI IV 536 | 9 (18%) | 21 (42%) | 43 (86%) | <0.001 |
| PAI II CFT073 | 3 (6%) | 7 (14%) | 19 (38%) | <0.001 |
| PAI I 536 | 1 (2%) | 3 (6%) | 18 (36%) | <0.001 |
| PAI II 536 | 0 (0%) | 2 (4%) | 11 (22%) | <0.001 |
| PAI I J96 | 0 (0%) | 0 (0%) | 2 (4%) | 0.329 |
| PAI II J96 | 2 (4%) | 5 (10%) | 15 (30%) | 0.001 |
| PAI I CFT073 | 7 (14%) | 13 (26%) | 37 (74%) | <0.001 |
| Adhesion genes | ||||
| afa | 1 (2%) | 5 (10%) | 3 (6%) | 0.298 |
| bmaE | 0 (0%) | 3 (6%) | 8 (16%) | 0.007 |
| fimH | 47 (94%) | 49 (98%) | 46 (92%) | 0.534 |
| gaf D | 0 (0%) | 1 (2%) | 10 (20%) | <0.001 |
| focG | 2 (4%) | 3 (6%) | 11 (22.0%) | 0.006 |
| Toxin genes | ||||
| cdtB | 3 (6%) | 5 (10%) | 9 (18%) | 0.194 |
| hlyD | 4 (8%) | 1 (2%) | 13 (26%) | 0.001 |
| cnf1 | 2 (4%) | 0 (0%) | 13 (26%) | <0.001 |
| cva C | 0 (0%) | 33 (66%) | 10 (20%) | <0.001 |
| usp | 4 (8%) | 3 (6%) | 27 (54%) | <0.001 |
| vat | 2 (4%) | 2 (4%) | 48 (96%) | <0.001 |
| sat | 1 (2%) | 4 (8%) | 22 (44%) | <0.001 |
| picU | 1 (2%) | 1 (2%) | 21 (42%) | <0.001 |
| Miscellaneous genes | ||||
| fliC H | 4 (8%) | 5 (10%) | 13 (26%) | 0.020 |
| ibeA | 10 (20%) | 13 (26%) | 7 (14%) | 0.325 |
| ompT | 29 (58%) | 29 (58%) | 31 (62%) | 0.895 |
| Protectins genes | ||||
| KPS MTI | 24 (48%) | 27 (54%) | 23 (46%) | 0.707 |
| KPS MT II | 15 (30%) | 23 (46%) | 35 (70%) | <0.001 |
| KPS MT III | 3 (6%) | 1 (2%) | 7 (14%) | 0.083 |
| rfc | 3 (6%) | 1 (2%) | 3 (6%) | 0.700 |
Table 1. Distribution of virulence factors and specific genes in fecal E. coli and urinary E. coli.
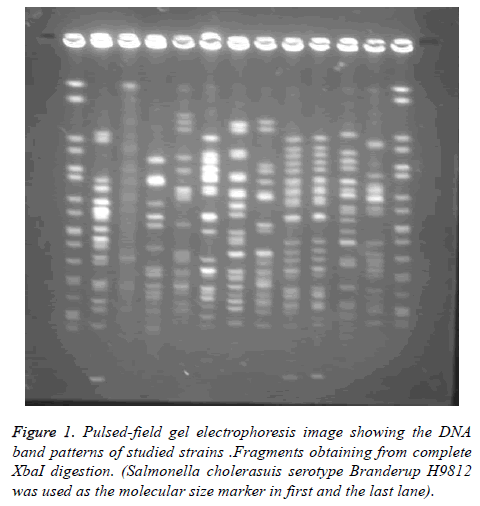
PFGE was performed in order to study the genetic linkage analysis among different sources of E. coli isolates. Genome sequence data showed that fragments obtaining from complete XbaI digestion would be anticipated only as 19 visible distinguishable fragments.
Of 150 strains examined in this study by pulsed field gel electrophoresis, 28 strains (18.6%) were identified as nontypeable. The numbers of bands obtained were varied considerably, and were overall out of 15 to 24 (mean 19 bands). The sizes of the bands were Kb 20-1135. Since some were colorless bands, Pierson’s similarity coefficient was used. The PFGE patterns of 150 distinct strains of E. coli were visually classified into pulsotypes with the assistance of BioNumerics software.
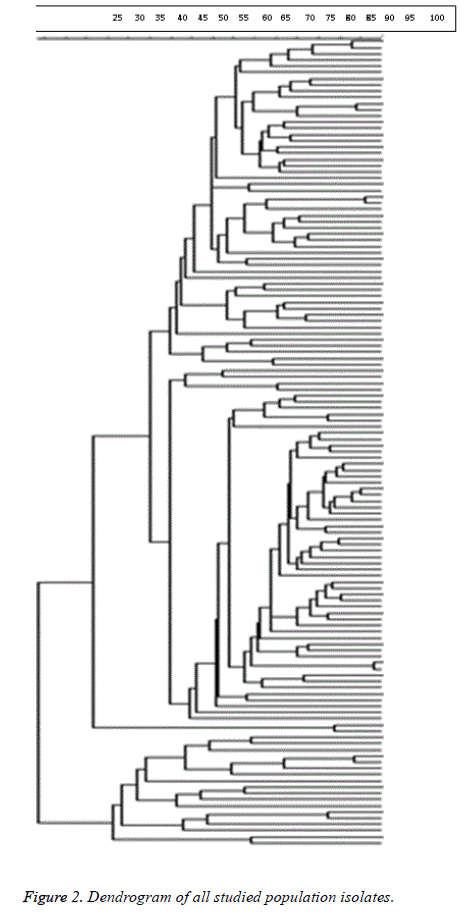
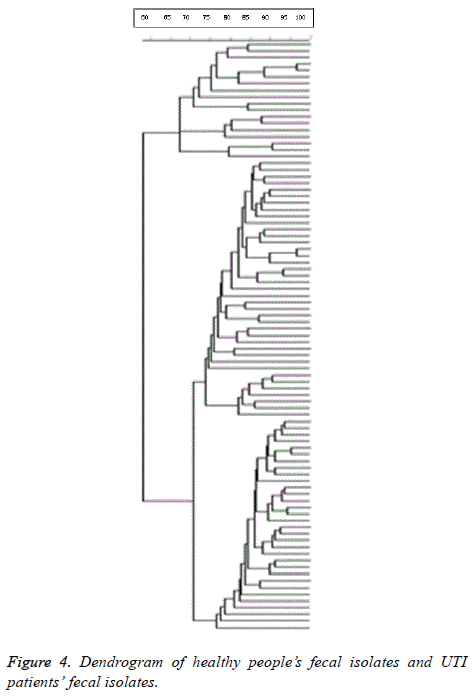
98% identity level and a similarity cut-off index of 76% were used to define PFGE profiles. Dendrogram drawing in the menu curve was conducted based on the peak bands. The high genetic diversity of DNA restriction pattern by means of PFGE was reported as following: 122 pulsotypes, including 14 clusters in all studied E. coli isolates; 84 pulsotypes with 14 clusters in fecal and urinary E. coli isolates derived from UTI patients; and 80 pulsotypes containing 8 clusters in fecal E. coli isolates derived from healthy people and UTI patients (Figure 1). Isolates with unrelated patterns were considered unrelated epidemiologically. The number of pulsotypes, clusters, and Dice coefficient based on studied isolates is shown in Table 2.
| Isolates | Pulsotypes (No.) | Clusters (No.) | Cluster (No.) | Dice coefficient |
|---|---|---|---|---|
| All studied isolates | 122 | 14 | Cluster1 (4), Cluster 2 (3), Cluster 3 (3), Cluster 4 (2), Cluster 5 (2), Cluster 6 (2), Cluster 7 (6), Cluster 8 (4), Cluster 9 (2), Cluster 10 (2), Cluster 11 (24), Cluster 12 (8), Cluster 13 (3), Cluster 14 (2) | 0.98 |
| Fecal and urinary isolates from UTI patients | 84 | 14 | Cluster 1 (6), Cluster 2 (15), Cluster 3 (2), Cluster 4 (2), Cluster 5 (2), Cluster 6 (2), Cluster 7 (2), Cluster 8 (2), Cluster 9 (5), Cluster 10 (2), Cluster 11 (3), Cluster 12 (3), Cluster 13 (2), Cluster 14 (1) | 0.912 |
| Fecal isolates from healthy people and UTI patient | 80 | 8 | Cluster 1 (7), Cluster 2 (2), Cluster 3 (4), Cluster 4 (3), Cluster 5 (27), Cluster 6 (2), Cluster 7 (7), Cluster 8 (22). | 0.908 |
Table 2. Number of pulsotypes, number of clusters and Dice coefficient according to studied isolates.
As shown in Figure 2, clonal spreading was not observed in the studied population due to high diversity profiles of PFGE. (Di=0.98) Dendrogram of all studied population isolates is provided below.
14 clusters were seen in fecal and urinary E. coli strains. Due to the high diversity profiles of PFGE, clonal spreading was not observed in fecal and urinary strains isolated from UTI patients. Dice coefficient of studied isolates was 0.912. Dendrogram of fecal and urinary strains isolated from UTI patients is shown in Figure 3.
8 clusters were identified. Due to the high diversity profiles of PFGE, clonal spreading was not observed in healthy people’s fecal isolates and UTI patients’ fecal isolates respectively. Dice coefficient of studied isolates was 0.908. Dendrogram of healthy people’s fecal isolates and UTI patients’ fecal isolates is shown in Figure 4.
Discussion
E. coli is an intestinal flora whose diversity creates large uncertainties in the identification of new genomes [20,21]. Most strains of E. coli are not harmful as long as they are living in the intestinal tract, but they may cause bacterial infections if they enter other parts of the body, such as conjunctiva, gallbladder, genital tract, eyes, appendix, and respiratory tract [22].
The development of PFGE typing methods based on fingerprinting of bacterial genome has given valuable tools to confirm the relationship among outbreak strains [23-27].
In the current study, we applied PFGE followed by XbaI restriction digestion of chromosomal DNA to determine the genetic relatedness among E. coli isolates derived from different sources. This enzyme is the most frequently used restriction enzyme for UPEC outbreaks [28,29]. The number of bands obtained was varied, and they were overall out of 15 to 24 (mean 19 bands). The sizes of the bands were 20 to 1135 kb. Since the samples were collected in a year and from different sources of population, these results were predictable. Only, 4 identical clones detected in all urinary and fecal isolated from patients with UTI. Two urinary and fecal strains isolated from 1 patient with UTI had more than 98% similarity. Also, four identical strains were reported in fecal isolates of healthy individuals (one strain belonged to a sister and her brother, and the other strain belonged to a twin .The best description of this variation was that the isolates from the subjects were not of a common clone or origin, and they did not have the same genotypes. Other factors that caused this difference including the number and size of the bands obtained from the fingerprinting .Also, these variations have been observed in different part of the world. Some reasons are being diverse of strains isolated from patients in distinct parts of the world, being various laboratory conditions, and a wide variety of E. coli strains in several human societies, and being genetic differences of strains because of different types of samples and their location. E. coli isolates belonging to the clonal group had different virulence characteristics. The probable reasons for the presence of the same virulence factors in pathogens and a large variety in the pulsotypes were being variety of E. coli in the colon and as a consequence of the diversity of the uropathogens. Other reason was possibility of genetic DNA exchange through horizontal gene transfer between strains.
Other studies have shown different bands with high diversity of molecular weights. In molecular typing of Japanese E. coli isolates, Kawamori et al. reported 19 to 24 DNA bands with molecular sizes of 30 to 500 kb With PFGE typing of UPEC strains, Ejrnaes et al. reported 15 to 20 distinct bands with 50 to 1200 kb molecular weights [30,31]. In an epidemiologic study of E. coli isolates in Northern Ireland, Watabe et al. observed 15 DNA fragments using PFGE [27].
In this study, the Dice similarity coefficient has been explained approximately as equal genetic relatedness [13,30,31], though there was little correlation among similarity evaluation obtained from single enzymes. Poor similarity and little correlation between Dice similarities can be explained by one enzyme digestion and by an enhanced pathogenic factor: the latter occurs due to the acquisition of virulence genes from the environment in bacterial genome. This hypothesis is supported by our finding. Based on other studies, single-enzyme digestion PFGE is presumably correlated deficiently with randomly amplified polymorphic DNA [13,32,33]. According to our study, there are some reasons for the weak correlations among Dice coefficients obtained from single fragment size resolution: these could explain the discrepancy between the 18-25 fragments identified by superlative published protocols for E. coli PFGE. On the other hand, concealed bands would not be considered and would influence the Dice index underneath. Two bands with simultaneous movement in a nonsame position could be considered as only one non-matching band and would influence the Dice coefficient upgrade [13,34,35].
To summarize, we can conclude that our results suggest that the presence of certain features such as PAI ICFT073, IICFT073, PAI I536, PAI IV536, PAI II J96, PAI II536, gafD, focG, vat, usp, hlyD, sat, cnf1, picU, fliC(H7), kpsMTII, kpsMTIII in E. coli has made the bacterium compatible with its extra-intestinal environment and may cause extra-intestinal infections in humans. On the other hand, the presence of some virulence genes such as PAI IV 536, fimH, cva C, ompT, KSP MT I, KSP MT II in the fecal bacteria can make it a reservoir for the emergence of virulence factors for ExPEC.
“Indeed, when an outbreak research is under way, time and resources may not incorporate more attempts. Without prior presumption permitted by epidemiologic data, however, a relatedness valid measure, rather than a method that simply recognizes matches, is required. Inferences about genetic linkages based on only one-enzyme PFGE should be mentioned with caution. The computer software programs analyse Dice coefficients and generate dendrograms relatively easily. In addition, failure to recognize relatedness concepts and similarity coefficients may have facilitated acceptance of dendrograms from single enzyme PFGE as precise relatedness measures. BioNumerics software produces banding patterns analysis from large sets of isolates, but it cannot be trusted to consider the presence or lack of bands without visual perception of the gel picture” [14,21,36]. The present study findings allow us to conclude epidemiologic relationships among isolates from PFGE data: however, more restriction enzymes would be required to make a reasonable approximation of genetic relatedness. Thus, poor resolution of fragment size could account for the discrepancy between the 18 to 25 fragments detected by most published PFGE protocols for O157 and the 40 fragments expected based on sequence data. There are other possible explanations for the discrepancy: (i) the sequence data may be in error or (ii) some of the XbaI recognition sites in the O157 genome may not be available to the enzyme if, for example, they happen to be adjacent to a methylated site. The impact of occult or comigrating bands is potentially great [37].
The observation of some virulence factors in urinary tract isolates, including: PAI ICFT073, IICFT073, PAI I536, PAI IV536, PAI II J96, PAI II536, gafD, focG, wat, usp, hlyD, sat, cnf1, picU, fliC (H7), kpsMTII, kpsMTIII compared to fecal isolates was statistically significant. The frequency of pathogenic factors in urine strains was significantly higher than fecal strains of patients and healthy individuals. Phylogenetic typing showed that urinary strains belong to the phylogenetic groups of typing B2 and D, while the fecal strains of the patients belonged to phylogenetic typing groups A and D and fecal isolates of healthy individuals belonged to phylogenetic typing groups A and B1. According to the findings of other studies, phylogenetic typing of B2 and D is most commonly seen in extracellular and pathogenic E. coli. The simultaneous presence of some virulence factors with high frequency in urinary strains compared to fecal isolates indicated that the pathogens producing UTI have a potential pathogenic compared to other isolates of this study. The differences between the distribution of virulence factors and genomic typing between urinary and fecal strains of patients with healthy individuals’ fecal isolates suggest that colonization of the virulent strain in the intestines can increase the risk of UTI.
Extra-intestinal pathogenic Escherichia coli (ExPEC) have a complex phylogeny, broad virulence factor (VF) and significant genomic plasticity, and are associated with a spectrum of host infective syndromes ranging from simple urinary tract infection to life-threatening bacteremia. Their importance as pathogens has come to the fore in recent years, particularly in the context of the global emergence of hypervirulent and antibiotic resistant strains. Despite this, the mechanisms underlying ExPEC transmission dynamics and clonal selection remain poorly understood. Large-scale epidemiological and clinical studies are urgently required to ascertain the mechanisms underlying these processes to enable the development of novel evidence-based preventative and therapeutic strategies [38].
Isolates derived from UTI patients did not have a common origin and are not identical genotypes. Differences in size and number of bands obtained from strains fingerprinting in different parts of the world, diverse experimental conditions in other parts of the world, a wide variety of E. coli in communities, and various human genetic variations due to several types of strains isolated from gathered samples cause these varieties.
Study Limitation
There are established methods such as DNA-seq (Illumina), Pacbio or Oxford Nanopore sequencing which they can easily assess the genetic diversity across the strains and probably discover some additional, new features. Due to the high cost of mentioned methods and limitations in our budget, we could not use them.
Recommendations
The present study provides molecular and epidemiological information about virulence factor genes found in two groups of E. coli. It is necessary to have a simple pathotypes screening test which can be beneficial to facilitate along with other experiments in establishing an UTI assessment. Unfortunately, due to the high variation in pathogenicity determinants of UPEC strains, pathotypes could not be determined using pathogenicity determinants. Knowledge of the molecular details of UPEC is mainstay of successful strategies development for treatment of UTI and prevention of its subsequent complications. Globally, PFGE method is capable of identifying 90% of the bacterial genome, and of monitoring genetic changes in antibiotic-resistant genes in hospitalinfection producing bacteria. Despite the high cost of materials and instruments, or the time-consuming procedure, PFGE is suggested as a practical and applicable typing method. Yet, it should be noted that although PFGE is taken as the goldstandard technique in epidemiological studies, using this method alone does not completely confirm the similarity of the two isolates. Thus, the utilization of the second strains typing method or additional epidemiological analysis is useful to understand the dominant gene and how genetic differences cause diseases.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Blango MG, Mulvey MA. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother 2010; 54: 1855-1863.
- Sabate M, Moreno E, Perez T, Andreu A, Prats G. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect 2006; 12: 880-886.
- Sivick Kelsey E, Mobley Harry LT. Waging war against uropathogenic Escherichia coli: winning back the urinary tract. Infect Immun 2010; 78: 568-585.
- Johnson JR, Scheutz F, Ulleryd P, Kuskowski MA, OBryan TT. Phylogenetic and pathotypic comparison of concurrent urine and rectal Escherichia coli isolates from men with febrile urinary tract infection. J Clin Microbiol 2005; 43: 3895-900.
- Katouli M. Population structure of gut Escherichia coli and its role in development of extra-intestinal infections. Iran J Microbiol 2010; 2: 59-72.
- Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A. Characterization of the Escherichia coli strain associated with an outbreak of hemolytic uremic syndrome in Germany: a microbiological study. Lancet Infect Dis 2011; 11: 671-676.
- Navidinia M, Najar Peerayeh S, Fallah F, Bakhshi B. Phylogenetic groups and pathogenicity island markers in Escherichia coli isolated from children. Jundishapur J Microbiol 2013; 6: e8362.
- Navidinia M, Najar Peerayeh S, Fallah F, Bakhshi B. Distribution of pathogenicity islands markers (PAIs) in uropathogenic E. coli isolated from children in Mofid Children Hopital. Arch Pediatr Infect Dis 2013; 1: 75-79.
- Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. Genomic anatomy of Escherichia coli O157: H7 outbreaks. Proc Nat Acad Sci USA 2011; 108: 20142-20147.
- Rumpa LV, Gonzalez-Escalonab N, Jua W, Wanga F, Caoa G. Genomic diversity and virulence profiles of historical Escherichia coli O157 strains isolated from clinical and environmental sources. Appl Env Microbiol 2015; 81: 569-577.
- Gholami Parizad E, Valizadeh A. The application of pulsed field gel electrophoresis in clinical studies. J Clin Diagn Res 2016; 10: 1-4.
- Schwartz D, Cantor C. Separation of yeast chromo some sized DNA by pulsed field gel electrophoresis. Cell 1984; 37: 67-75.
- Davis MA, Hancock DD, Besser TE, Call DR. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157: H7. J Clin Microbiol 2003; 41: 1843-1849.
- Sabat AJ, Budimir A, Nashev D, Sa-Leao R, Dijl Jm V. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eur Surveill 2013; 18: 20380.
- Goering RV. Pulsed field gel electrophoresis: a review of application and interpretation in the molecular epidemiology of infectious disease. Infect Gene Evol 2010; 10: 866-875.
- Standard Operating Procedure for Pulse Net PFGE of Escherichia coli O157: H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. PNL05 2013; 1-13.
- Najar Peerayeh S, Navidinia M, Fallah F, Bakhshi B, Jamali J. Pathogenicity determinants and epidemiology of uropathogenic E. coli (UPEC) strains isolated from children with urinary tract infection (UTI) to define distinct pathotypes. Biomed Res 2018; 29: 2035-2043.
- Carriço JA, Pinto FR, Simas C, Nunes S, Sousa NG. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J Microbiol 2005; 43: 5483-5490.
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995; 33: 2233-2239.
- Hosseini M, Anvarinejad M, Mohammadzadegam R. Microbial susceptibility, virulence factors, and plasmid profiles of uropathogenic Escherichia coli strains isolated from children in Jahrom, Iran. Arch Iran Med 2012; 15: 312-316.
- Zhang L, Foxman B, Manning SD, Tallman P, Marrs CF. Molecular epidemiologic approaches to urinary tract infection gene discovery in uropathogenic Escherichia coli. Infect Immun 2000; 68: 2009-2015.
- Sandt C, Krouse D, Cook C, Hackman A, Chmielecki W. The key role of pulsed- field gel electrophoresis in investigation of a large multiserotype and multistate food-borne outbreak of Salmonella infections centered in Pennsylvania. J Clin Microbiol 2006; 44: 3208-3212.
- Alfizah H, Nordiah AJ, Rozaidi WS. Using pulsed field gel electrophoresis in the molecular investigation of an outbreak of Serratia marcescnes infection in an intensive care unit. Singapore Med J 2004; 45: 214-218.
- Derakhshan S, Najar-Peerayeh S, Bakhshi B. Multiple-locus variable number tandem repeat analysis of Klebsiella pneumoniae: comparison with pulsed-field gel electrophoresis. Microb Drug Resist 2017; 23.
- Najar Peerayeh S, Eslami M, Memariani M, Siadat SD. High prevalence of blaCTX-M-1 group extended-spectrum β-lactamase genes in Escherichia coli isolates from Tehran. Jundishapur J Microbiol 2013; 6: 6863.
- Najar Peerayeh S, Rostami E, Eslami M, Ahangarzadeh Rezaee M. High frequency of extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Escherichia coli isolates from male patients urine. Arch Clin Infect Dis 2016; 11: 32696.
- Watabe M, Hogg GM, Millar BC, Crothers L, Rooney PJ. Epidemiological study of E. coli 0157: H7 isolated in Northern Ireland using pulse field gel electrophoresis. Ulster Med J 2008; 77: 168-174.
- Basim H. Pulsed field gel electrophories technique and its use in molecular biology. Turkish J Biol 2000; 25: 408-418.
- Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R. The optimization of a rapid pulsed field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Japan J Infect Dis 2009; 62: 372-377.
- Ejrnaes K, Sandvang D, Lundgren B, Ferry S, Holm S. Pulsed field gel electrophoresis typing of Escherichia coli strains from samples collected before and after pivmecillinam or placebo treatment of uncomplicated community acquired urinary tract infection in women. J Clin Microbiol 2006; 44: 1776-1781.
- Radu S, Ling OW, Rusul G, Karim MIA, Nishibuchi M.Detection of Escherichia coli O157: H7 by multiplex PCR and their characterization by plasmid profiling, antimicrobial resistance, RAPD, and PFGE analyses. J Microbiol Meth 2001; 46: 131-139.
- Kawamori F, Hiroi M, Harada T, Ohata K, Sugiyama K. Molecular typing of Japanese Escherichia coli 0157: H7 isolates from clinical specimens by multilocus variable number tandem repeat analysis and PFGE. J Med Microbiol 2008; 57: 58-63.
- Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 2014; 9: 104984.
- Doran G, Morris D, OHare C, DeLappe N, Bradshaw B, Corbett-Feeney G. Cost-effective application of pulsed-field gel electrophoresis to typing of Salmonella enterica Serovar Typhimurium. Appl Env Microbiol 2005; 71: 8236-8240.
- Kah Heng S, Kek Heng C, Puthucheary SD. Stacking gels: a method for maximising output for pulsefield gel electrophoresis. Ind J Med Microbiol 2009; 27: 142-145.
- Kudva IT, Davis MA, Griffin RW, Garren J, Murray M, John M. Polymorphic amplified typing sequences and pulsed-field gel electrophoresis yield comparable results in the strain typing of a diverse set of Bovine Escherichia coli O157: H7 isolates. Int J Microbiol 2012.
- Davis MA, Hancock DD, Besser TE, Call DR. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157: H7. J Clin Microbiol 2003; 41: 1843-1849.
- Dale AP, Woodford N. Extra-intestinal pathogenic Escherichia coli (ExPEC): Disease, carriage and clones. J Infect 2015; 71: 615-e626.