Research Article - Journal of RNA and Genomics (2019) Volume 15, Issue 1
Detection of phage and in-silico analysis of WO phage associated cif genes from Wolbachia: A study based on Drosophila model
Kopal Singhal and Sujata Mohanty*Department of Biotechnology, Jaypee Institute of Information Technology, Noida, India
- *Corresponding Author:
- Mohanty S
E-mail: sujata.mohanty@jiit.ac.in, Fax: 0120-2400986, Tel: +91-1202594208
Received date: 11 February 2019; Accepted date: 23 February 2019; Published date: 02 March 2019
Abstract
Wolbachia are endosymbiotic bacteria infecting the arthropod and nematode population inducing various reproductive manipulations in the host to enhance their transmission. 90% of the arthropod infecting Wolbachia is known to possess a bacteriophage named WO phage. Together the WO phage, Wolbachia and host form a unique tripartite association. Bacteriophages contain mobile genetic elements and play a crucial role in horizontal gene transfer. Also, WO phage is known to assist Wolbachia in inducing Cytoplasmic incompatibility (CI) in the host, as the two cytoplasmic incompatibility factor genes i.e., CifA and CifB are present in the eukaryotic association model of the WO phage. In the present study, we detected the presence of WO phage in Indian Drosophila host from five different eco-geographic locations using the Orf7 gene marker and found their presence to be limited to the Wolbachia infecting host. The multiple sequence alignment of Orf7 gene from two Wolbachia strains reveals a conserved sequence throughout. We also compared the Cif genes across six Wolbachia genomes and found interesting strain-specific variations. These single nucleotide differences in Cif genes can be explored functionally to understand their role in inducing CI. This can also explain the variations in the CI levels across the different Wolbachia strains. The present study is a preliminary work to understand phage distribution and Cif genes across the Indian Wolbachia genomes. Functional validation of the key findings in the study can help in establishing Wolbachia as a robust model for vector borne disease control.
Keywords
WO phage; Wolbachia; Drosophila tripartite; Cytoplasmic incompatibility; Cif genes
List of Abbreviations
CI: Cytoplasmic Incompatibility; Cif: Cytoplasmic Incompatibility factor ; wMel_Ref: Wolbachia endosymbiont of Drosophila melanogaster from NCBI database; wMel_Kl: Wolbachia endosymbiont of Drosophila melanogaster from Kerala, India ; wMel_AMD: Wolbachia endosymbiont of Drosophila melanogaster from Ahmedabad, India; wRi_Ref: Wolbachia endosymbiont of Drosophila simulans from NCBI database; wRi_KL: Wolbachia endosymbiont of Drosophila ananassae from Kerala,India; wRi_AMD: Wolbachia endosymbiont of Drosophila ananassae from Ahmedabad,India; wPip: Wolbachia pipentis strain; D. melanogaster: Drosophila melanogaster; D. ananassae: Drosophila ananassae; SNV: Single Nucleotide Variation
Introduction
Bacteriophages are the viral particles associated with bacterial genomes. One such bacteriophage the WO Phage, infect the endosymbionts Wolbachia [1]. Phage-Wolbachia-host forms a unique tripartite association wherein phage resides inside the Wolbachia strain which itself is in symbiosis with the host. Literature reports the restricted presence of WO phage in arthropod host and its absence from nematode host, where Wolbachia shares a mutualistic association [2]. Phylogenetic analysis carried out to understand the evolutionary history of WO phage with the Wolbachia host points towards an independent evolution of phage genome from the host genome [1,3,4]. Phage genomes possess the ability to carry out horizontal gene transfer and this ability can be exploited in the use of Wolbachia strains as a biocontrol agent for prevention of vector-borne diseases [5,6].
Studies have linked the presence of WO phage to the various reproductive manipulations exhibited in the arthropod host such as Cytoplasmic Incompatibility (CI) [7]. WO phage carries some signature domains e.g. ankyrin repeats whose role in CI has also been hypothesised [7,8]. CI can be either unidirectional or bidirectional and results from mating of Wolbachia uninfected female with infected male giving rise to unviable progeny, however, the infected female rescues the host from CI by making the progeny viable for both infected and uninfected [9]. CI incompatible crosses results in defects in paternal chromosomal condensation and separation leading to embryonic lethality. Wolbachia is known to be associated with the reproductive tissues of the host but the mature sperm of the infected male lacks Wolbachia and will produce viable progeny only if the egg is uninfected or carries the same Wolbachia strain [10]. However, the detailed underlying mechanism is still not clear [11]. Level of CI and its penetrance is dependent on host- Wolbachia-phage interactions [7]. While wMel strain is reported to show zero to low levels of CI in D. melanogaster, wRi strain shows high CI levels in D. simulans [12]. Also when wMel is transfected into D. simulans high CI levels were observed highlighting the host specific nature of CI. Two Wolbachia genes identified in the eukaryotic association model (WO phage genes encoding proteins having eukaryotic functions) in WO phage were reported for their role in CI [13]. Further, these genes were named as Cif (Cytoplasmic incompatibility factor). The functional role of CifA and CifB in causing CI was elucidated in some studies and contrasting results were observed. While transgenic expression analysis using the Cif homologs of wPip reveal inability of these genes in rescuing CI, the analysis from wMel Cif genes show that CifA alone as well as in association with CifB rescues host from CI, whereas, CifB gene alone was unable to rescue CI [14].
The tripartite association of phage-Wolbachia-host is intriguing and yet remains unexplored. Role of WO phage in cytoplasmic incompatibility which itself is a tool of Wolbachia persistence inside the host is challenging and needs deeper insight. In the present work, we detected the presence of WO phage in five natural populations of Drosophila host and performed a comparative analysis of the Cif genes linked with the eukaryotic association model of phage.
Materials and Methods
PCR based phage detection and Sanger sequencing across Drosophila host
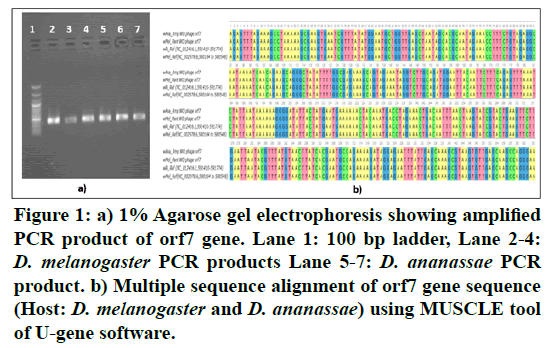
PCR based detection for WO phage was done in Drosophila flies collected from 5 different regions (Rampur, Bhubaneswar, Pantnagar, Jabalpur, Delhi) of India using Orf7 gene marker at 57°C (Tm) from gDNA of host fly [15]. The quality of both gDNA as well as PCR product was checked using gel electrophoresis and the samples were run on 1% agarose gel. Two PCR products showing positive results for phage (one each from Drosophila melanogaster and Drosophila ananassae) with maximum band intensity and quality were purified using ExoSAP cleaning protocol (Thermo Scientific). The cleaned PCR products were sequenced using Sanger sequencing and the results were edited and analysed using DNAstar software. Multiple sequence alignment using MUSCLE tool of U-gene software was performed taking two Indian gene sequences (wAna_orf7, wMel_orf7) and two reference sequence i.e., wRi_Ref (NC_012416.1,591410-591774), wMel_Ref (NC_002978.6,580184-580548) of Orf7 gene [16,17].
Identification of CifA and CifB gene across 6 Wolbachia genomes
Four whole genomes of two Wolbachia strains from Drosophila host generated earlier were utilised in the present study [18]. CifA (Gene ID: 29555381) and CifB (Gene ID: 34927827) gene sequences of Wolbachia endosymbiont of D. melanogaster (wMel_Ref) were retrieved from NCBI database. These sequences were blast against four whole genomes of Indian Wolbachia (Table 1) as well as the reference Wolbachia wRi genome and the corresponding sequences in these genomes were extracted.
| Genome (Accession number) | CifB (Locus) | Number of genes predicted | Gene (info) | CifA (Locus) | Number of genes predicted | Genes info |
|---|---|---|---|---|---|---|
| wRi_KL MKIF00000000 | 506692 to 510213 | 1 | 212_aa|+|1|639 | 505213 to 506637 | 1 | 474_aa|+|1|1425 |
| 2 | 945_aa|+|685|3522 | |||||
| wRi_AMD MSYL00000000 | 458825 to 462345 | 1 | 203_aa|+|1|612 | 457346 to 458770 | 1 | 474_aa|+|1|1425 |
| 2 | 945_aa|+|684|3521 | |||||
| wRi_Ref NC_012416.1 | 573202 to 576723 1079661 to 1083182 | 1 | 212_aa|+|1|639 | 571723 to 573147 1078182 to 1079606 | 1 | 474_aa|+|1|1425 474_aa|+|1|1425 |
| 2 | 945_aa|+|685|3522 | 1 | ||||
| wMel_KL MLZJ00000000 | 532600 to 536121 | 1 | 1173_aa|+|1|3522 | 536176 to 537600 | 1 | 474_aa|+|1|1425 |
| wMel_AMD MNCG00000000 |
539234 to 542755 | 1 | 1173_aa|+|1|3522 | 537755 to 539179 | 1 | 474_aa|+|1|1425 |
| wMel_Ref NC_002978 |
618702 to 622223 | 1 | 1173_aa|+|1|3522 | 617223 to 618647 | 1 | 474_aa|+|1|1425 |
Table 1: Blastn identified locus and genemarks based gene predictionof Cif genes in the studied Wolbachia genomes.
Comparative genomics and structural analysis of Cif genes
Multiple sequence alignment for the 6 CifA and CifB gene sequences was performed using Muscle tool of U-gene software [16,17]. Gene prediction for these nucleotide sequences was done using GenemarkS [19]. The protein sequences provided by GenemarkS were used to predict the protein structural domains with HHpred using the default parameters and databases SCOPe70 (v.2.06), Pfam (v.31.0), SMART (v6.0), and COG/ KOG (v1.0) [13,20].
Results and Discussion
Molecular basis of phage identification
The Wolbachia WO capsid protein Orf7 used as a detection marker for phage revealed that the presence of phage was limited to Wolbachia infected Drosophila hosts i.e., Phage was found to be present only in D. melanogaster and D. ananassae from five eco-geographical locations of India in Figure 1A. An earlier study on Indian Drosophila also proposed that phage is an integral part of the Wolbachia genome and was found to be absent from Wolbachia uninfected Drosophila [15].
Figure 1: a) 1% Agarose gel electrophoresis showing amplified PCR product of orf7 gene. Lane 1: 100 bp ladder, Lane 2-4: D. melanogaster PCR products Lane 5-7: D. ananassae PCR product. b) Multiple sequence alignment of orf7 gene sequence (Host: D. melanogaster and D. ananassae) using MUSCLE tool of U-gene software.
The MUSCLE tool alignment for these four Orf7 genes sequences produced a 365 bp position alignment and revealed a conserved gene sequence throughout the length in Figure 1B. Earlier studies done to understand the dynamics of WO phage with their corresponding Wolbachia host reveal incongruence with respect to Wolbachia phylogeny [3,4,6]. These results highlighted the fact that presence of a specific phage is not linked to the functional effects depicted by Wolbachia in the host. However, this observation raises a fundamental question on the persistence of WO phage in the Wolbachia genomes.
Cif genes: Comparative and structural analysis
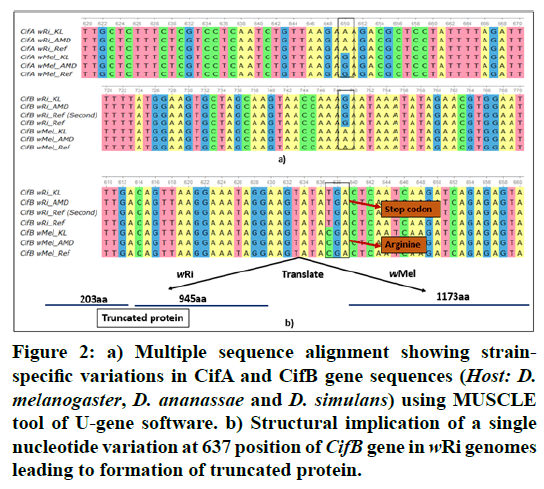
In the present study, we identified CifA and CifB genes in the six Wolbachia genomes using Blast searches. A single copy of both CifA and CifB gene was found to be present in all studied Wolbachia genomes with the exception of wRi_Ref, where these genes were found to be duplicated as seen in Table 1. In all cases, the CifA gene was located upstream of CifB. Literature reports that Cif genes were found to be associated with the eukaryotic association model of prophage [21]. Although, it has been noticed earlier that the presence of phage in Wolbachia genomes may not be an indication of the reproductive phenotypes induced by Wolbachia, but the role of Cif genes in causing Cytoplasmic Incompatibility in the host is recently documented [13,22,23]. The multiple sequence alignment produced a 1425bp position alignment for CifA and 3522bp alignment for CifB gene and reveal strain-specific variations in Figure 2A. GenemarkS tool used for gene prediction revealed the presence of a complete copy CifA protein sequence (474 aa) in all Wolbachia genomes, however, in case of CifB a complete protein sequence (1173 aa) was limited to the wMel strain as seen in Table 1. In case of wRi strains a single site substitution (TC) at 637 positions in the nucleotide sequence resulted in a change of codon from CGA (Arginine) to TGA which is a stop codon in Figure 2B. This SNV (single nucleotide variation) resulted in a truncated protein sequence with two regions predicted in CifB of wRi, each of approximately 200 and 900 amino acids. Similar results were observed in an earlier study for wRi_Ref [22]. This single nucleotide variation may have direct functional implication in the wRi genomes which are even known to rescue the bidirectional Cytoplasmic Incompatibility caused by wMel strain [12,22].
Figure 2: a) Multiple sequence alignment showing strainspecific variations in CifA and CifB gene sequences (Host: D. melanogaster, D. ananassae and D. simulans) using MUSCLE tool of U-gene software. b) Structural implication of a single nucleotide variation at 637 position of CifB gene in wRi genomes leading to formation of truncated protein.
Cif genes have been categorised into four variants i.e., Type I, II, III and IV on the basis of structural analysis [13,22]. As per literature, Type I variants of Cif genes were found in both wMel and wRi genomes and a Type II variant was reported only for wRi_Ref [13,22]. Similarly, in our study, Type I variants were present in both wRi and wMel Indian genomes however, the Type II variant of the Cif genes was not identified in Indian wRi genomes. We subjected our protein sequences to HHpred server for identification of functional domains in the proteins as explained in Table 2. One catalase and apoptosis regulatory domain was predicted in CifA gene, however, due to poor matching scores no significant functions could be assigned for these domains. CifB gene was found to possess a conserved peptidase domain possessing proteolytic activity as seen in Table 2. In case of wRi genomes, the truncated protein sequence 1 of 200 aa was found to possess regulatory domains such as RNA promoter binding domain or transcription factors but due to low matching scores the exact functions of this region could not be stated. The other 900 aa sequence also possess the peptidase domain. An earlier study has also reported the presence of a putative catalase domain in the CifA region and predicted its function in preventing from the damage done by oxidative stress [14]. The role of CifB was derived from a sequence homolog in wPip strain i.e., CidB which has a deubiquitylating capacity and a cysteine protease active site [24]. Transgenic studies to understand the role of these cid genes in yeast and Drosophila model were also carried out. However, the knowledge about these genes is fairly new and needs further experimental validations. The existence of WO phage in 90% of the arthropod infecting Wolbachia and location of Cif genes in the flanking regions of the WO phage raises the possibility of a direct association between these CI inducing genes and WO phage.
| Gene ID | Name | Function | Probability | E-value |
|---|---|---|---|---|
| wRi_KL: CifA | Bcl-2_3 family protein | Apoptosis regulatory protein | 54.33 | 16 |
| DUF249 ; Multigene family 530 protein | Domain of Unknown function | 29.94 | 530 | |
| NST1 | Salt tolerance down-regulator | 26.85 | 170 | |
| Catalase-rel | Catalase-related immune-response | 21.31 | 280 | |
| wMel_KL: CifA | Bcl-2_3 | Apoptosis regulatory protein | 54.65 | 16 |
| DUF249 ; Multigene family 530 protein | Domain of Unknown function | 33.09 | 440 | |
| NST1 | Salt tolerance down-regulator | 31.52 | 140 | |
| Catalase-rel | Catalase-related immune-response | 21.2 | 280 | |
| wRi_KL: CifB | PDDEXK_9 ; PD-(D/E)XK nuclease superfamily | Hypothetical bacterial proteins | 98.07 | 1.00E-07 |
| Ulp1 protease C-terminal domain | Ubiquitin like specific protease | 97.32 | 0.0000045 | |
| Peptidase_C48 ; Ulp1 protease family, C-terminal catalytic domain | proteolytic activity | 95.93 | 0.00094 | |
| wMel_KL: CifB | PDDEXK_9 ; PD-(D/E)XK nuclease superfamily | Hypothetical bacterial proteins | 97.85 | 4.80E-07 |
| Ulp1 protease C-terminal domain | Ubiquitin like specific protease | 96.84 | 0.000069 | |
| Peptidase_C48 ; Ulp1 protease family, C-terminal catalytic domain | proteolytic activity | 94.96 | 0.013 |
Table 2: Hhpred based protein functional domain prediction in the Cif genes of studied Wolbachia genomes.
Conclusion
The present work raises several interesting questions on the tripartite association of phage-Wolbachia-host. It compels the researchers to think whether the strain-specific variations in Cif genes is responsible for the differences in reproductive manipulation by Wolbachia. Do the non-CI inducing strains possess Cif homologs? In addition, wMel shows weak CI, whereas, wRi shows high CI levels. So if wRi Cif genes are transfected into wMel strain it can be made more virulent for the vector control programme. An understanding of all the significant genes like Ankyrin, type iv secretion system, Cif genes and their functional role can help scientist to create a superstrain of Wolbachia capable of controlling the spread of various arboviral diseases nd malaria, at the same time maintaining its persistence in the host species. Experimental studies designed to bridge this gap can prove to be a boon in making Wolbachia an ideal vector borne disease control agent.
Acknowledgments
The authors thank Biolinkk India Pvt Ltd. for providing sanger sequencing service. KS thanks CSIR for funding the sequencing expense and providing SRF-Fellowship. The authors also thank the Vice Chancellor, JIIT for providing infra-structure facilities for conducting this work.
Competing Interest
Authors declare no competing interest.
References
- Masui S, Kamoda S, Sasaki T, et al. 2000. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol 51, 491–497.
- Kent BN and Bordenstein SR. 2010. Phage WO of Wolbachia: Lambda of the endosymbiont world. Trends Microbiol 18, 173–181.
- Gavotte L, Henri H, Stouthamer R, et al. 2006. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol 24, 427–435.
- Bordenstein SR and Wernegreen JJ. 2004. Bacteriophage flux in endosymbionts (Wolbachia): Infection frequency, lateral transfer, and recombination rates. Mol biol evol 21, 1981–191.
- Tanaka K, Furukawa S, Nikoh N, et al. 2009. Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integration into the Wolbachia genome. Appl Environ Microbiol 75, 5676–5686.
- Wang GH, Sun BF, Xiong TL, et al. 2016. Bacteriophage WO can mediate horizontal gene transfer in endosymbiotic wolbachia genomes. Front Microbiol 7, 1867.
- Bordenstein SR, Marshall ML, Fry AJ, et al. 2006. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog 2, e43.
- Singhal K and Mohanty S. Genome organisation and Comparative genomics of four novel wolbachia genome assemblies from Indian drosophila host. Funct Integr Genomics.
- Telschow A, Flor M, Kobayashi Y, et al. 2007. Wolbachia-induced unidirectional cytoplasmic incompatibility and speciation: Mainland-island model. PLoS One 2, e701.
- Veneti Z, Clark ME, Zabalou S, et al. 2003. Cytoplasmic incompatibility and sperm cyst infection in different drosophila-wolbachia associations. Genetics 164, 545–552.
- Serbus LR, Casper LC, Landmann F, et al. 2008. The genetics and cell biology of wolbachia host interactions. Annu Rev Genet 42, 683–707.
- Poinsot D, Bourtzis K, Markakis G, et al. 1998. Wolbachia transfer from drosophila melanogaster into D. simulans: Host effect and cytoplasmic incompatibility relationships. Genetics 150, 227–237.
- Lindsey AR, Rice DW, Bordenstein SR, et al. 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol Evol 10, 434–451.
- Shropshire JD, On J, Layton EM, et al. 2018. One prophage WO gene rescues cytoplasmic incompatibility in drosophila melanogaster. Proc Natl Acad Sci 115, 4987–4991.
- Ravikumar H, Prakash BM, Sampathkumar S, et al. 2011. Molecular subgrouping of wolbachia and bacteriophage WO infection among some Indian drosophila species. J Genet 90, 507–510.
- Edgar RC. 2004 MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113.
- Golosova O, Henderson R, Vaskin Y, et al. 2014. Unipro UGENE NGS pipelines and components for variant calling, RNA-seq and ChIP-seq data analyses. Peer J 2, e644.
- Singhal K and Mohanty S. 2018. Comparative genomics reveals the presence of putative toxin–antitoxin system in Wolbachia genomes. Mol Genet Genom 293, 525–540.
- Besemer J, Lomsadze A, Borodovsky M, et al. 2001. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29, 2607–2618.
- Soding J, Biegert A, Lupas AN, et al. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic acids res 33, W244–W248.
- Bordenstein SR and Bordenstein SR. 2016. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun 7, 13155.
- LePage DP, Metcalf JA, Bordenstein SR, et al. 2017. Prophage WO genes recapitulate and enhance wolbachia-induced cytoplasmic incompatibility. Nature 543, 243.
- Asselin AK, Villegas OS, Hoffmann AA, et al. 2018. Contrasting patterns of virus protection and functional incompatibility genes in two conspecific Wolbachia strains from Drosophila pandora. Appl Environ Microbiol AEM, 02290.
- Beckmann JF, Ronau JA, Hochstrasser M, et al. 2017. A wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol 2, 17007.