- Biomedical Research (2015) Volume 26, Issue 2
Comparative Prevalence of Community-Acquired-Methicillin-resistant Staphyloccocus aureus (CA-MRSA) among Students of Centro Escolar University (Philippines), Kumamoto Health Science University (Japan) and Daegu Health College (Korea)
Chikateru Nozaki1, Takayuki Masaki1, Su Jung Kim4, Rogelio S. Cruz2, Charito M. Bermido2, Kyung-Yong Kim3, Cheolin Park4*1Faculty of Health Science, Kumamoto Health Science University, Japan.
2College of Medical Technology, Centro Escolar University, Philippines.
3Department of Social Welfare, Daegu Health College, Korea.
4Department of Biomedical Laboratory Science, Daegu Health College, Korea
- *Corresponding Author:
- Cheolin PARK
Department of Biomedical Laboratory Science
Daegu Health College
15 Youngsong-Ro, Buk-gu, Daegu, Korea
Accepted date: December 07 2014
Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) has been recognized as a nosocomial pathogen since the 1960s. Cases of MRSA infection presenting from the community were first described in the early 1980s and there had been a growing concern in the development of Methicillin- Resistant Staphylococcus aureus (MRSA). Nowadays, the increase in incidence of CA-MRSA was reported and was recognized as a threat by health professionals. Generally, CA-MRSA strains carry the Panton-Valentine leukocidin (PVL) virulence genes, which is a bicomponent leukocidin and causes leukocyte destruction, tissue necrosis and possess a mobile staphylococcal cassette chromosome mec (SSCmec). Based on this serious health concern, the prevalence of CAMRSA with other parameters such as pvl and SSCmec from different three communities (countries) was investigated. The subjects in this collaborative research were 200 students of CEU, 100 of DHC, and 94 of KHSU. Stapylococcal cassette chromosome mec and Panton-Valentine leukocidin gene were investigated using a multiplex PCR. The prevalence of MRSA from CEU, DHC and KHSU observed was, 12%, 3% and 3.2%, respectively. In the case of PVL, it was found in 3% of the samples from CEU, but was not detected in any of the samples of DHC and KHSU. SSCmec was not always carried by CA-MRSA strains. This collaborative study is the first trial of its kind among the three communities representing the three countries, South Korea, Japan and the Philippines. The information elicited will help understand the prevalence of CAMRSA.
Keywords
Staphylococcal cassette chromosome mec (SSCmec), Panton-Valentine leukocidin (PVL), communityassociated methicillin-resistant Staphylococcus aureus MRSA (CA-MRSA)
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has been recognized as a nosocomial pathogen since the 1960s. Cases of MRSA infection presenting from the community were first described in the early 1980s [1]. MRSA is a potentially fatal strain of Staphylococcus aureus that is resistant to several antibiotics including oxacillin. MRSA is categorized based on how it is acquired. The first type, healthcareacquired MRSA (HA-MRSA) were identified in patients in hospital and healthcare facilities that were resistant to methicillin. The more serious concern is a second type of MRSA which appeared in the 1990s and is known as communityacquired MRSA (CA-MRSA). CA-MRSA occurs outside of hospitals and usually infects the skin of an otherwise healthy individual. Recently, this bacterium was found to account for an increasing amount of infections acquired among wrestling players, students, and soldiers. Unlike HA-MRSA, the source of infection of CA-MRSA is often difficult to identify [2].
CA-MRSA usually differs from several ways. Typically, they carry the smallest staphylococcal cassette chromosome mec (SCCmec) types IV and V, are resistant to fewer antimicrobial agents, and are associated with the presence and enhanced expression of specific virulence factors [2,3]. Indeed, Panton-Valentine Leukocidin (PVL) has been associated with severe and complicated CA-MRSA osteoarticular infections. PVL causes leukocyte destruction and necrotizing pneumonia, an aggressive condition that is often fatal within 72 hours. Comparing cases of staphylococcal necrotizing pneumonia, 85% of community- acquired cases were PVL-positive, while none of the hospital-acquired cases were found positive. It has played a role in a number of outbreaks of fatal bacterial infections [4-6]. The aim of this study is to determine the prevalence of CA-MRSA using several parameters such as mecA, PVL gene as well as SCCmec from CA-MRSA isolates from college and university students from the three different three countries, South Korea, Japan and the Philippines.
Materials and Methods
For this collaborative study, three parameters, CA-MRSA, mecA gene and PVL gene were used in the investigation but there were a little modification depends on each community.
DHC researched and collected the samples from the nasal cavities and hands of 100 college students. KHSU performed the research on samples from nasal vestibule and the back of ear auricle of 94 students. CEU collected samples from nares/noses and palms of 100 college students.
i) From DHC
Using sterile cotton swabs, the isolates were obtained and transferred into non-selective and later selective medium, for enrichment and selection, respectively. This was followed by incubation for 24hrs, DHC performed using VITEK (BioMerieux, Maryl’Etoile, France)-automated system, to identify and to test for the antimicrobial susceptibility of the bacteria. The standard bacteria, Staphylococcus aureus (ATCC 29213), Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27858) were used. DHC adapted the multiplex PCR assay previously published paper [4] for mecA and PVL primers. The SCCmec primers sets were employed and determined on the basis of the band pattern obtained [7]. The ethical committee of DHC permits for conducting this study, and all study participants were provided with informed and written consent.
ii) From KHSU
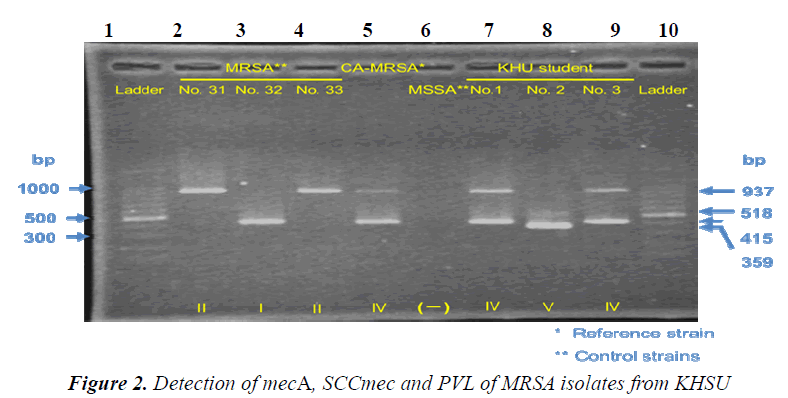
Strains. CA-MRSA strains (SCCmec genotype IV, and PVL gene Positive) were provided as reference strains from Kyushu University Hospital.Control strains (No. 31: SCCmec genotype II /PVL gene Negative, No. 32: SCCmec genotype I/PVL gene Negative, No. 33: SCCmec genotype II/PVL gene Negative、 MSSA: mecA Negative/PVL gene Negative) were provided from Kumamoto University Hospital. No.1 strain, No.2 strain, and No.3 strain were isolated from Nasal vestibule and the back of ear auricle of our students using screening medium. All procedures were approved by the institutional review board of Kumamoto KHSU.
Determination of CA-MRSA
The most commonly known carrier of the mecA gene is the bacterium known as MRSA, which encodes the protein PBP2A (Penicillin binding protein 2A). With this fact, KHSU performed for the detection of PBP2’, which was examined by MRSA Latex Test (DENKA SEIKEN CO., INC.). The mecA gene and Panton-Valentine leukocidin gene (PVL) were detected [8] and typing of SCCmec was performed [7]. Drug susceptibility testing was performed using the method of Microscan WalkAway (SIMENS).
iii) From CEU
The study was carried out from September to November 2013 in Centro Escolar University, Centennial laboratory. A cross sectional study was conducted on a total of 100 students from ten different colleges. Collections of the nasal and palm samples from the participants were approved by the CEU- Institutional Ethics Committee. Nasal and palm swabs were collected and transferred in the Microbiology Laboratory. Sterile cotton swabs were used for sample collection. The sample was obtained by rotating the swabs gently on anterior nasal. Using another sterile cotton swab, samples were collected on both palmar surface of the palms of the participants and placed in the Nutrient broth and incubated overnight at 37°C. All catalase and coagulase positive Gram positive cocci isolates were confirmed using VITEX 2 XL for biochemical and antimicrobial profiles. However, CEU adapted a single PCR assay [9].
Results
1. Antimicrobial susceptibility test
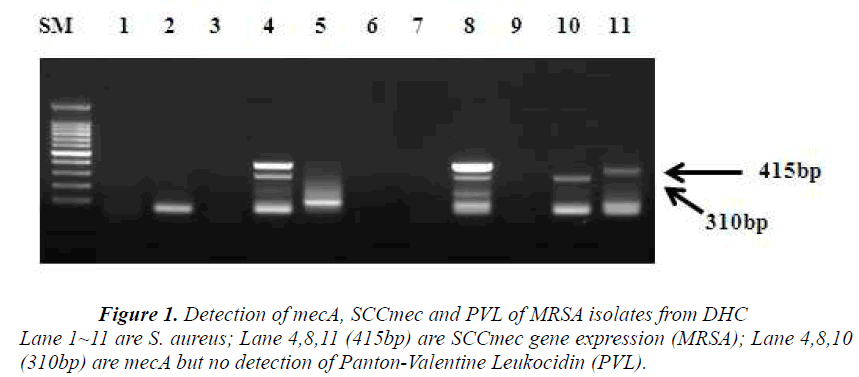
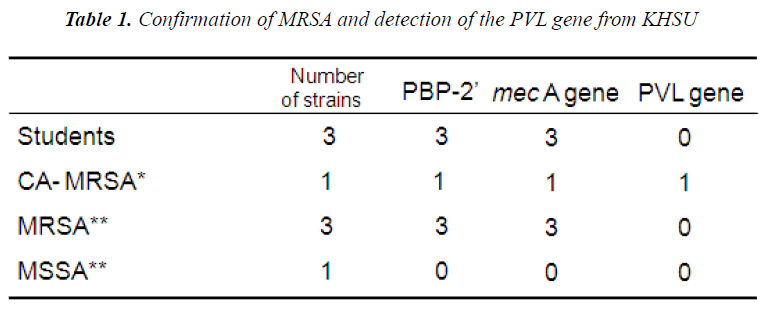
The CEU study revealed that 12% (14 nares/nose and 10 palm swabs /200 samples) were resistant to the antimicrobial, oxacillin. Among 24 samples, 5 strains were chosen as MRSA based on its resistance to cloxacillin. The study of DHC revealed that 15 strains were identified and 11 candidates from the group were selected to be MRSA, the ones which had shown <90% identification rate with VITEK-automated system. These are presumptive MRSA strains.. However, out of 11 isolates, only 3 strains (27%) were identified as MRSA as they carry mecA gene (Figure 1). KHSU had found out that 3 samples (3.2%) from 94 students were identified as MRSA by Microscan Walk- Way (Table 1). From antimicrobial susceptibility test, CEU has about 4 times higher on oxacillin resistant rate those of DHC and KHSU as MRSA rate.
The most commonly known carrier of the mecA gene is the bacterium known as MRSA, which encodes the protein PBP2A (Penicillin binding protein 2A). With this fact, KHSU performed the test to detect PBP2’. Three isolates out of 94 students were identified as MRSA and all of them were shown to carry PBP2’ which confirms them as MRSA on the protein level.
2. Detection of mecA, SSCmec and PVL genes
Following the antimicrobial susceptibility test, DHC detected PVL gene, SCCmec and mecA gene from 11 presumptive MRSA strains, using a multiplex PCR. Three isolates were confirmed as MRSA since they carried mecA gene and SCCmec. From three isolates, two samples have these two parameters, simultaneously. However, PVL gene was not detected (Figure 1).
Results of Penicillin Binding Protein 2’ reflected those of mec gene. All the isolated strains harbored no PVL gene. Strain No.1, No2 and No.3 are SCCmec IV, SCCmec V and SCCmec IV, respectively. Drug susceptibility testing showed that the three strains were sensitive to CEZ、IPM、and MINO, and that one strain was sensitive but the others were resistant to EM and CLDM. Our results similar to those by previously published reports [10,11], suggesting that AC-MRSA have spread within the community of Kumamoto in Japan.
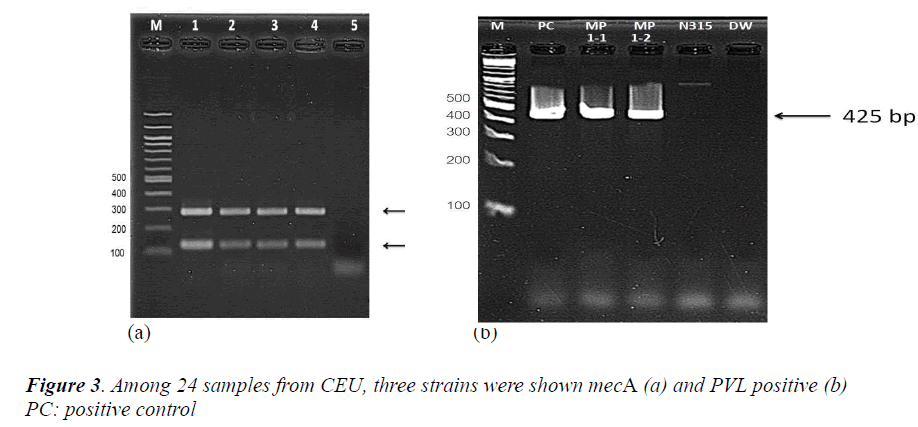
From an antimicrobial susceptibility test, CEU screened 24 samples as presumptive MRSA and finally chose 5 samples to be confirmed. The identification of S. aureus (femA) and MRSA (mecA) were carried out with mecA of 286 base pairs (Figure 3-a). CEU detected the PVL gene from two colonies of a single sample and performed same with other two samples. The samples which carry mecA gene were also detected PVL gene (Figure 3-b, other two samples were same so not shown here).
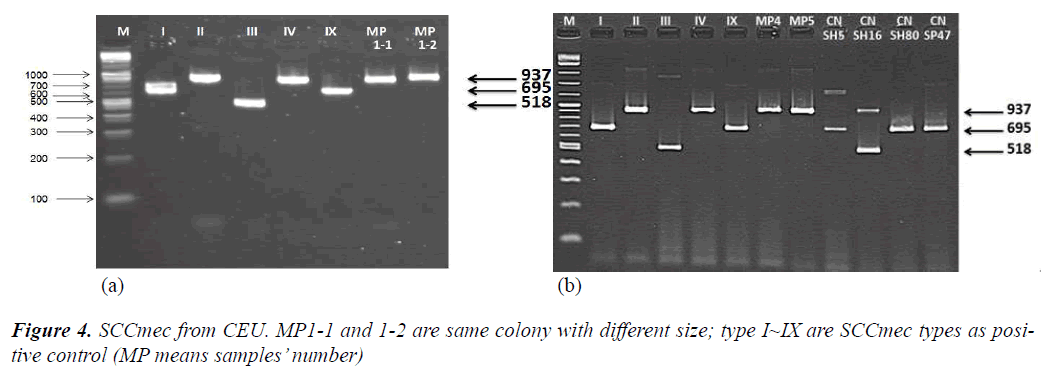
From another single PCR to detect SCCmec and to determine SCCmec type, CEU observed that all three samples were shown as SCCmec type IV (Figure 4-a,b).
Discussion
During the past years methicillin resistant S. aureus (MRSA) has emerged as the community acquired methicillin resistant S. aureus (CA-MRSA) [7]. Among three different communities like colleges and universities, the prevalence rate of CA-MRSA was shown to be high (12%) in the Philippines, and low (3.2% and 3.0%) in the Japan and Korea, respectively. The rate of MRSA in all community-associated S aureus infections in Asian countries ranges from 2·5% to 39% [12]. CA-MRSA can be spread by close skin-to-skin contact, touching contaminated stuffs and surfaces, unsanitary and crowded living conditions and poor personal hygiene. These are the usual modes of transmission of CA-MRSA. The slightly higher prevalence rate of CA-MRSA in CEU could be caused by humid and crowded conditions. Most of CA-MRSA strains have the virulence factor, Panton-Valentine Leukocidin (PVL), which is not frequently found in hospitalacquired MRSA strains or methicillin-sensitive S. aureus (MSSA) strains but a low incidence of MSSA was reported, and has been associated with necrotizing pneumonia and death [12].
The most commonly known carrier of the mecA gene is the bacterium known as MRSA. The detection of PVL and methicillin resistance (mecA) genes represents a new tool to aid the early identification of CA-MRSA isolates [4]. Panton-Valentine leukocidin is a biocomponent leukocidin encoded by two cotranscribed genes, lukS-PV and lukF-PV (lukS/F-PV), which causes leukocyte destruction and tissue necrosis [4]. PVL is also an S. aureus-specific exotoxin and its genes have been demonstrated primarily among CA-MRSA [2]. Several studies has shown that PVL genes were found <5% of S. aureus isolates worldwide [5,12,13], whereas our collaborative study showed three samples from CEU were PVL positive, however, the PVL genes were not detected in samples from KHSU and DHC. The strain carrying mecA gene has PVL genes and considered very important factor for virulence because the PVL toxin has been shown to be responsible for many of the severe clinical symptoms of infection caused by MRSA, such as furunculosis, severe necrotizing pneumonia, and necrotic lesions of the skin and soft tissues.
However, the rates of carriage of the PVL toxin gene are >75% for CA-MRSA stains [14] and 67% and 80% for CA-MRSA strains with ST8 and ST1, respectively, in the United State [15]. This big different rate of PVL positive from CA-MRSA might be caused by bacterial strains and direct isolates from healthy human skin of this study. Consequently, the reported articles were studied with MRSA bacterial strains, which are presumptively carrying PVL gene. It is expected that CA-MRSA infection is associated with community PVL-positive MRSA from nasal cavity and skin, but not detected from CA-MRSA. The detection of PVL and methicillin resistance (mecA) genes represents a new tool to aid the early identification of CAMRSA isolates [4]. Using multiplex PCR, both the two parameters, PVL and mecA were not found in the 10 samples but separately detected. The identification of S. aureus isolates carrying PVL genes is an important first step in controlling the virulence [4]. The combination of both PVL and mecA shows very potent virulence and resistant to antibiotics such as oxacillin.
However, it was expected that mecA and PVL genes are simultaneously expressed but not correlated from DHC and KHSU. Consequently, it is very important to identify serious and harmful S. aureus from both detection of PVL and mecA. However, several studies [4] showed these patterns PVL(-) MSSA, PVL(+) MSSA, PVL (-) MRSA and PVL (+) MRSA. Therefore, the PVL toxin can be carried by both MRSA (methicillin resistant Staphylococcus aureus) and MSSA (methicillin sensitive Staphylococcus aureus). The PVL genes are more common in CA-MRSA isolates compared with CA-MSSA isolates [14-16] and a recent molecular study showed the presence of PVL gene in almost all of the CA-MRSA strains [17].
The resistance of bacteria to antimicrobials is caused by the acquisition of the methicillin resistance gene mecA, carried by the staphylococcal cassette chromosome mec (SCCmec). By the time the Guidelines for the classification of SCCmec elements was prepared, eight SCCmec types have been described for S. aureus [20]. SCCmec types IV and V elements are generally carried by CAMRSA strain as reported [21,22]. Therefore, it is expected that samples 4,8 and 11 of DHC which has mecA gene can be also carried by SCCmec but samples 4 and 8 were observed using the two parameters, mecA and SSCmec. A previous paper [7] reported two-type-size of primers of SCCmec and DHC designed and adapted primers of SCCmec type IV with 415 bp size. Therefore, samples 4 and 8 were regarded as SCCmec type IV. SCCmec type IV is the most common type, followed by type V, type I and type II [7]. CA-MRSA generally carries mecA gene with PVL-positive and SCCmec but it can be expected that MSSA also can carry SSCmec. KHSU also got two SCCmec type IV with 415bp and one SCCmec type V with 359 bp. In the case of CEU, all of three samples carrying mecA gene could be expressed by SCCmec type IV. However, it was reported that CA-MRSA isolates carried the SCCmec type IV complex, and most were PVL positive, whereas the HA-MRSA isolates carried the SCCmec type II complex and did not harbor the PVL genes [2,23]. The two parameters, mecA and SCCmec were negative in the samples from DHC and three samples KHSU. However, CEU confirmed three samples which have mecA, SCCmec and PVL gene together. The mecA gene encodes for an altered penicillin binding protein known as penicillin binding protein 2A and thereby endows resistance to methicillin and other similar beta-lactam antimicrobial agents. Resistance to methicillin by staphylococcus results from the acquisition of the mecA gene, which is part of a larger mobile genetic element known as the staphylococcal chromosomal cassette (SCCmec), a genetic element that integrates into the staphylococcal chromosome. PVL is present in the majority of community acquired methicillin resistant S. aureus and is produced from the genetic material of a bacteriophage which infects S. aureus, making it more virulent [24]. Therefore, we can generalize that, “MRSA strain has PBP2A encoded by mecA which is located on a SCCmec element and are sometimes carried by PVL gene”.
The study was performed on college students taking health science courses, 200 from CEU, 100 from DHC and 94 from KHSU. It was shown that there is no high rate of PVL-positive mecA and SCCmec. PVL-positive isolates is more serious but CA-MRSA do always carry PVL gene. This proves that the higher rate of prevalence of the three parameters, PVL-positive, mecA and SCCmec can be acquired through exposure to unsanitary environments. From the results of this first collaborative study on the comparison of prevalence rates in the three different communities, information to elucidate on the three parameters, mecA, SCCmec and PVL gene from CA-MRSA is made available and can be useful to future researchers who wish to probe deeper on MRSA. Furthermore, sampling, raw data collection be standardized to elicit reliable results / information with the use of other molecular diagnostic tools. The result of this study is not conclusive but is vital and priceless in the area of community-acquired MRSA of the three different countries, South Korea, Japan and the Philippines.
Acknowledgement
We thank Dr. Patcharee Jearanaikoon and Dr.Aroonlug Lulitanond of the Faculty of Allied Medical Sciences, Khon Kaen University,Khon Kaen, Thailand, who provide their priceless comments and assistance.
References
- Frazee BW, Lynn J, Charlebois ED, Lambert L, Lowery D, Perdreau-Remington F. High Prevalence of Methicillin-Resistant Staphylococcus aureus in Emergency Department Skin and Soft Tissue Infections Annals of Emergency Medicine 2005; 45(3):311-320.
- Moroney SM, Heller LC, Arbuckle J, Talavera M, Widen RH Staphylococcal Cassette Chromosome mec and Panton-Valentine Leukocidin characterization of Methicillin-Resistant Staphylococcus aureus clones. J Clin Microbiol 2007; 45(3); 1019-1021.
- Vandenesch, F, Naimi, T, Enright,M. C, Lina, G, Nimmo, G. R, Heffernan, H, Liassine, N, Bes, M., Greenland, T. Community-acquired methicillin resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 2003; 9: 978-984.
- McClure J, Conly JM, Lau V, Elsayed S, Louie T, Huchins W, Zhang K. Novel Multiplex PCR assay detection of the Staphylococcal Virulence Marker Panton-Valentine Leukocidin genes and stimultaneous discrimination of methicillin-susceptible from-resistant Staphylococci. J Clin Microbiol 2006; 44(3): 1141-1144.
- Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999; 29: 1128-1132.
- Zhang K, J. Sparling, B. L. Chow, S. Elsayed, Z. Hussain, D. L. Church, D. B. Gregson, T. Louie, and J. M. Conly. 2004. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol 2004; 42: 4947-4955.
- Boye, K., Bartels, M.D., Andersen, I.S., Møller, J.A. and Westh, H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I–V. Clin Microbiol Infect 2007; 13: 725-727.
- Lina G, Piémont Y, Florence G, Bes M Peter M, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine Leukocidin—Producing Staphylococcus aureus in Primary Skin Infect Pneumonia. 1999; 29: 1128-1132.
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of Multiplex PCRs for Staphylococcal Cassette Chromosome mec Type Assignment: Rapid Identification System for mec, ccr, and Major Differences in Junkyard Regions. Antimicrob Agents Chemother 2007: 51(1); 264-274.
- Shamsudin M, Sekawi Z, Belkum A, Neela V. First community-acquired meticillin-resistant Staphylococcus aureus in Malaysia. Med Microbiol 2008; 57(9): 1180-1181.
- Ma XX, Sun DD, Wang S, Wang ML, Li M, Shang H, Wang EH, Luo EJ. Nasal carriage of methicillinresistant Staphylococcus aureus among preclinical medical students: epidemiologic and molecular characteristics of methicillin-resistant S. aureus clones, Diagnostic Microbio Infect. Disease. 2011; 70(1): 22-30.
- Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. Communityacquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 2003; 9: 978-984.
- Gillet, Y., B. Issartel, P. Vanhems, J. C. Fournet, G. Lina, M. Bes, F. Vandenesch, Y. Piemont, N. Brousse, D. Floret, and J. Etienne. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002; 359: 753-759.
- Hamilton, S. M., A. E. Bryant, K. C. Carroll, V. Lockary, Y. Ma, E. McIndoo, L. G. Miller, F. Perdreau-Remington, J. Pullman, G. F. Risi, D. B. Salmi, and D. L. Stevens. In vitro production of Panton-Valentine leukocidin among strains of methicillin-resistant Staphylococcus aureus causing diverse infections. Clin Infect Dis 2007; 45: 1550-1558.
- McAleese, F., E. Murphy, T. Babinchak, G. Singh, B. Said-Salim, B. Kreiswirth, P. Dunman, J. O’Connell, S. J. Projan, and P. A. Bradford.Use of ribotyping to retrospectively identify methicillin-resistant Staphylococcus aureus isolates from phase 3 clinical trials for tigecycline that are genotypically related to communityassociated isolates. Antimicrob Agents Chemother 2005; 49: 4521-4529.
- Martinez-Aguilar G, Avalos-Mishaan A, Hulten K, HammermanW, Mason EO Jr, Kaplan SL. Communityacquired, methicillin-resistant and methicillinsusceptible Staphylococcus aureus musculoskeletal infections in children. Pediatr Infect Dis J 2004; 23: 701-706.
- Ellis MW, Hospenthal DR, Dooley DP, Gray PJ, Murray CK. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin Infect Dis 2004; 39: 971-979.
- Diep BA, Sensabaugh GF, Somboona NS, Carleton HA, Perdreau-Remington F. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clin Microbiol 2004; 42: 2080-2084.
- Avalos Mishaan AM, Mason EO Jr, Martinez-Aguilar G. Emergence of a predominant clone of communityacquired Staphylococcus aureus among children in Houston, Texas. Pediatr Infect Dis J 2005; 24: 201-206.
- Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009; 53(12): p. 4961-4967.
- Boyle-Vavra, S. & Daum, R. S. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest 2007; 87: 3-9.
- Tristan A, Michele Bes, Meugnier H, Lina G, Bozdogan B,Courvalin P, Reverdy M, Enright MC, Vandenesch F, Etienne J Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis 2007; 13: 594-600.
- Shannon M. Moroney, Loree C. Heller, Jesse Arbuckle, Monica Talavera and Ray H. Widen Staphylococcal Cassette Chromosome mec and Panton-Valentine Leukocidin Characterization of Methicillin-Resistant Staphylococcus aureus Clones J Clin Microbiol 2007; 45(3): 1019-1021.
- Voyich JM, Otto M, Mathema B. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis. 2007; 194(12): 1761-1770.