Review Paper - Journal of Genetics and Molecular Biology (2021) Volume 5, Issue 2
ARX gene with an impressive role in X-linked intellectual disability
1Department of Medical Genetics, School of Medicine, University of Medical Sciences, Tehran
2Human Genetics Research Center, Baqiyatallah University of Medical Sciences, Tehran
- *Corresponding Author:
- Shirin Ghadami
Department of Medical Genetics
School of Medicine
University of Medical Sciences
Tehran, Iran
Tel: + (98 21) 8898 9487
E-mail: shiringhadami@gmail.com
Accepted on February 01, 2019
Citation: Ghadami S, Eshaghkhani Y. ARX gene with an impressive role in X-linked intellectual disability. J Genet Mol Biol. 2019;3(1):1-10.
DOI: 10.35841/genetics-molecular-biology.3.5-14
Visit for more related articles at Journal of Genetics and Molecular BiologyAbstract
Intellectual disability is the most common neurodevelopmental defect in the worldwide. X-linked intellectual disability (XLID) is the frequent form of intellectual disability which includes a heterogeneous group of inherited disorders emerging as various degrees of intellectual disabilities. XLID has a prevalence of 2.6 cases per 1,000 in the general population and accounts for over 10% of all cases of intellectual disability. Based on associated phenotypes, XLID is subdivided into syndromic (S-XLID) and non-syndromic (NS-XLID) forms; where two third of XLID cases are thought to be non-syndromic. Among the non-syndromic form, the aristalessrelated homeobox gene (ARX) gene is one of the ideal candidates to be evaluated in NS-XLID, since its mutations are responsible for about 9.5% of XLID cases. The ARX is located on the Xp22.13 genomic region and encodes a highly conserved protein with a considerable role in Wnt/β-catenin signaling pathway. Base on review literature, mutations in ARX gene has a particular influence on the critical processes associated with the brain development. Our results in bioinformatics study of molecular features, second and quaternary structures of ARX gene and also the phylogeny tree of ARX protein is showed that the ARX is a highly conserved protein with a substantial role in an important developmental pathway and its deficiency can cause irreversible defects, mainly in brain, that leads to the development of XLID as a common form of intellectual disability and also, the sequence alignment of this protein with other spices confirms that the functional domains of ARX protein are highly conserved, thus it has been predicted that the mutations of this gene is highly pathogenic. Alongside, we mainly focused to gather the data addressing the structural properties of ARX protein and bioinformatics assay of this protein to find the important role of ARX gene in the integrity of normal brain development.
Keywords
ARX, Intellectual disability, X-linked, Structure, Wnt/β-catenin signaling.
Introduction
Intellectual disability (ID) is the most frequent neurodevelopmental disorder in the world characterized by an intelligence quotient (IQ) below 70. ID is associated with deficits in adaptive behaviours (including personal or social manners) before the age of 18 [1]. The prevalence of ID is approximately (2-3)% in the general population; however, it is widely different among populations based on epidemiological, cultural and nutritional backgrounds, and particularly for mild cases, it varies significantly based on several heterogeneous factors causing ID [2-8]. In addition to genetic factors, nongenetic (so-called environmental) susceptibility factors can also play a substantial role in increasing risk of ID. For example, prenatal exposure to teratogens (e.g., alcohol, infectious) and serious birth injuries are two main environmental factors which may participate in the pathogenesis of ID [9,10].
ID is divided into mild to moderate (IQ 50-70) and severe to profound (IQ<50) forms. Although the prevalence of severe form of ID is roughly stable, the prevalence of mild ID is unstable and specifically depends on the cultural and social demographic variables [1].
Literature Review
In a study, the prevalence of mild, moderate, severe and profound ID has been reported about 85%, 10%, 4%, and 2% of the all affected individuals, respectively [11]. ID or associated phenotypes resulted from a monogenic defect are subdivided into 4 categories according to the mode of inheritance, autosomal dominant ID, autosomal recessive ID, X-linked ID and mitochondrial ID [12-14]. Mutations in X-linked genes account for (5-10)% of all types of ID and are the most likely causes of ID in males [15].
X-linked Intellectual Disability (XLID)
X-linked Intellectual disability (XLID) includes a heterogeneous group of inherited disorders emerging as various degrees of intellectual disabilities, which is developed by mutations in genes located on the X-chromosome. XLID has a prevalence of 2.6 cases per 1,000 in the general population and accounts for over 10% of all cases of intellectual disabilities [16].
It was first on 1938 that Lionel Penrose observed a higher male: female ratio of intellectual disability in British population. This ratio was 1.25:1 [17]. The observation was further confirmed by numerous studies in different centers where they all agreed with the fact that there is an excess of 30% in affected males with ID. This tendency of men being more affected than females together with findings obtained from large mentally retarded pedigrees that show X-linked inheritance, raised the idea that this excess might be attributed to X chromosome [18-22]. Identification of fragile X syndrome in late 1970s was an important finding to approve this hypothesis [23]. Since then, several other X-linked IDs were described.
Based on associated phenotypes, XLID is subdivided into syndromic (S-XLID) and non-syndromic (NS-XLID) forms; where two third of XLID cases are thought to be non-syndromic [12,24]. In the case of S-XLID, more than 100 of the underlying loci or genes have been identified until now, while for the NS-XLID, this number does not exceed 50. Nevertheless, with the current advances in molecular genetics, this number is drastically increasing [13,25]. About 140 S-XLID have been descried so far among which Fragile X-syndrome is the most common cause of XLID [26]. It is estimated that 40% of the protein-coding genes identified on the human X chromosome are expressed in the brain. XLMR could result from mutations in any of these loci. In a recent update of genes involved in XLID, more than 100 X-linked genes have been identified where some of them play more important roles than the others [13,25].
Genes involved in XLID
Two genes playing considerable roles in syndromic XLID are FMR1 and FMR2. FMR1, the first XLID gene that cloned, encodes an mRNA-binding protein that acts during mRNA transport and translational regulation. Mutations in FMR1 lead to a unique phenotype called Fragile X syndrome (FXS). FXS is the most common form of XLID [23]. Expansions of CGG repeats (>200 repeats) in 5'UTR of FMR1 is the cause of FXS [27-30].
FMR2 is located distal to FMR1 and is separated only by LOC100128690, LOC100132556 and MIRN514-2 at Xq27.3 [31]. It has been thought that the screening of FMR2 gene mutations in patients that are negative for FMR1 mutation is essential. However, expansion repeats in this gene are relatively rare. FMR2 is a transcriptional regulator, and is probably involved in long-term memory and enhanced long-term potentiating [32-34].
Despite the growing number of genes identified in XLID, current information about their function is scarce. The major families of NS-XLID genes are involved in transcriptional regulation, cell proliferation, cell differentiation and cell locomotion (actin cytoskeleton), especially in central nervous system [35,36]. For instance, Rho proteins are highly conserved regulators of the actin cytoskeleton, cell adhesion and migration, cytokinesis and gene expression; hence, a number of genes contributing to XLID pathogenesis should be involved in Rho-GTPase signaling pathway. In humans, four XLID genes that participate directly in cellular signaling through Rho-GTPases are oligophrenin 1 (OPHIN1), PAK3, αPIX (ARHGEF6) and GD1 [37-40]. Oligophrenin acts as a Rho-GAP (GTPase-activating protein) and stimulates the GTPase activity of RHOA, RAC1 and CDC42 [38,41]. PAK3 is a serine/threonine protein kinase that accelerates the effects of downstream RAC1 and CDC42 on the actin cytoskeleton and gene expression [42]. in addition, αPIX is a guanine nucleotide exchange factor for RAC1 and CDC42 [39]; and GDI1 is involved in RHO-GTPase signaling through regulation of Rab4 and Rab5 pools, thus it is probably involved in the maturation of synaptic vesicles [43].
The selected list of involved XLID genes and their related phenotypes are described in Tables 1 and 2 [1]. ARX (ARX gene, OMIM no.300382, GenBank no.NM_139058) is the most frequent cause of intellectual disability after fragile X syndrome; and its mutations are highly associated with syndromic and non-syndromic forms of XLID [44], which will be discussed below in this review.
| Gene Mutated | Associated Disorders | Main Characteristics | Functions of Related Protein |
|---|---|---|---|
| ARX | X-linked mental retardation/ Early infantile epileptic encephalopathy-1/ X-linked lissencephaly/ Proud syndrome/ Partington syndrome | Moderate to profound mental defect/ Frequent tonic seizures or spasticity beginning in infancy, neurodevelopmental regression, dystonia /Lissencephaly, agenesis of the corpus callosum, structural brain anomalies, early-onset intractable seizures, severe psychomotor retardation, ambiguous genitalia/ Agenesis of the corpus callosum, severe mental defect, seizures, spasticity, microcephaly, urogenital anomalies/ Focal dystonia | Formation of dendritic spine, maintenance of special neuronal subtypes in cerebral cortex and axonal direction in the floor plate; neuronal proliferation and differentiation of GABA-releasing neurons |
| ATP6AP2 | X-linked Parkinson disease with spasticity/ X-linked mental retardation (Hedera type) | Parkinsonian features and variably penetrant spasticity/ Mild to moderate mental defect, epilepsy | Renin receptor; ERK1 and ERK2 activation |
| BCOR | Syndromicmicrophthalmia-2 | Microphthalmia, congenital cataract, dental abnormalities, urogenital and skeletal anomalies | Co-repressor for transcription; possible role in modulation of histone acetylation and chromatin remodeling |
| CDKL5 | Early infantile epileptic encephalopathy-2 | Infantile spasticity, seizures in the first months of life, severe global developmental delay resulting in mental defect and poor motor control, lack of speech development, subtle dysmorphic facial features, sleep disturbances, gastrointestinal problems, stereotypic hand movements | Serine-threonine kinase; chromatin remodelling |
| FGD1 | X-linked mental retardation/ Aarskog–Scott syndrome† |
Mental defect/short stature, hypertelorism, shawl scrotum, brachydactyly | RhoGEF; Possible role in stimulation of actin polymerization and neurite outgrowth |
| FLNA | Periventricular heterotopia/Otopalato-digital syndrome I-IV (including Melnick-Needles syndrome) | Epilepsy, cleft palate, short stature, skeletal and facial anomalies | Actin-binding protein; neurite outgrowth; formation of dendritic spine |
| FMR1 | Fragile X Syndrome | moderate to severe mental defect, macroorchidism, distinct facial features (including long face, large ears and prominent jaw) | mRNA-binding protein; mRNA transport and regulation of translation |
| GK | Glycerol kinase deficiency† | Hyperglycerolaemia, severe developmental delay, short stature, osteoporosis, recurrent spasticity | Nuclear translocation of the glucocorticoid-receptor complex |
| JARIDIC | X-linked mental retardation (Claes-Jensen type) | Microcephaly, recurrent spasticity, epilepsy, short stature, facial anomalies, mental defect | Transcription factor; chromatin remodeling |
| KIAA1202 | Stocco dos Santos X-linked mental retardation syndrome | Congenital hip dislocation, recurrent infections, short stature | PDZ domain-containing protein; possible role in actin remodeling |
| MAOA | Brunner syndrome/ Antisocial behavior | Impulsive aggressiveness and mild mental retardation/ Violent, hostile and tempestuous manners | Serotonin metabolism |
| MECP2 | Rett syndrome/X-linked mental retardation (Lubs type) | Neurodevelopmental regression, epilepsy, hand stereotypies, recurrent spasticity, recurrent spasticity/ mental defect | Silencing of transcription in neuronal genes |
| OPHN1 | X-linked mental retardation with cerebellar hypoplasia and distinctive facial appearance | Neonatal hypotonia with motor delay, Marked strabismus, Early-onset complex partial seizures, Moderate to severe mental defect | Negative control of rhoGTPases; stabilization of dendritic arbours |
| PHF6 | Borjeson-Forssman-Lehmann syndrome | Severe mental defect, epilepsy, hypogonadism, hypometabolism, obesity, swelling of subcutaneous tissue of face, narrow palpebral fissure, large but not deformed ears | PHD zinc-finger protein; putative role in transcription |
| PHF8 | X-linked mental retardation (Siderius type) | Mild to borderline mental defect, cleft lip or palate | PHD zinc-finger protein; putative role in transcription |
| PQBP1 | Renpenning syndrome | Microcephaly, short stature, small testes, dysmorphic face, ocular colobomas, cardiac malformations, cleft palate | Polyglutamine-binding; mRNA splicing |
| RSK2 | X-linked mental retardation-19/ Coffin–Lowry syndrome |
Mild to moderate mental defect/ skeletal malformations, growth retardation, hearing deficit, paroxysmal movement disorders, cognitive impairment | Serine-threonine protein kinase; CREB phosphorylation; long-term memory |
| SLC16A2 | Allan-Herndon-Dudley syndrome | Hypotonia, severe mental retardation, dysarthria, ataxia, athetoid movements, muscle hypoplasia, spastic paraplegia with hyperreflexia, clonus, Babinski reflexes, abnormal thyroid tests | Monocarboxylate transporter; transport of T3 into the cytoplasm |
| SLC6A8 | Cerebral creatine deficiency syndrome 1 | Epilepsy, mental retardation, severe speech delay, behavioral abnormalities, seizures, facial anomalies | Creatine transporter, maintenance of creatine pool in brain |
| SMS | Snyder–Robinson syndrome | Facial asymmetry, marfanoid habitus, unsteady gait, thickened lower lip, nasal dysarthic speech, narrow or cleft palate, diminished muscle mass, osteoporosis, kyphoscoliosis, long great toes, short stature, pectuscarinatum, myopia | Spermine synthase |
| SYN1 | X-linked epilepsy with variable learning disabilities and behavior disorders† | Macrocephaly, epilepsy, aggression | Synaptic-vesicle-associated protein |
| XNP | X-linked epilepsy, with variable learning disabilities and behavior disorders | Epilepsy, learning difficulties, macrocephaly, aggressive behavior, skeletal, urogenital and facial anomalles, α-thalassemia, short stature, spastic diplegia | DNA helicase; chromatin remodeling, DNA methylation and regulation of gene expression; regulator of cortical size |
Not always associated with intellectual disability. GABA: γ-amino butyric acid; ERK: Extracellular-Signal-Regulated Kinase; GEF: Guanine-Nucleotide Exchange Factor; MAO: Monoamine Oxidase; CREB: cAMP-Response-Element-Binding Protein; T3: Thyroid Hormone; ATR-X: X-Linked α-Thalassemia Mental Retardation.
Table 1. Genes responsible for syndromic form of X-linked intellectual disability.
| Gene Mutated | Associated Disorders | Main Characteristics | Functions of Related Protein |
| ACSL4 | X-linked mental retardation-63 | Nonspecific mental defect | Long-chain fatty-acid synthase, possible role in membrane synthesis/recycling |
| AGTR2 | X-linked mental retardation | Severe to profound mental defect | Brain-expressed angiotensin receptor 2 |
| ARHGEF6 | X-linked mental retardation-46 | Severe mental defect | Integrin-mediated activation of Rac and cdc42, stimulation of neurite outgrowth |
| ARX* | X-linked mental retardation/ Early infantile epileptic encephalopathy-1/ X-linked lissencephaly/ Proud syndrome/ Partington syndrome | Moderate to profound mental defect/ Frequent tonic seizures or spasticity beginning in infancy, neurodevelopmental regression, dystonia /Lissencephaly, agenesis of the corpus callosum, structural brain anomalies, early-onset intractable seizures, severe psychomotor retardation, ambiguous genitalia/ Agenesis of the corpus callosum, severe mental defect, seizures, spasticity, microcephaly, urogenital anomalies/ Focal dystonia | Formation of dendritic spine, maintenance of special neuronal subtypes in cerebral cortex and axonal direction in the floor plate; neuronal proliferation and differentiation of GABA-releasing neurons |
| DLG3 | X-linked mental retardation-90 | Moderate to severe mental defect | Post-synaptic scaffolding protein; linked to NMDA-type glutamatergic receptors |
| FGD1* | X-linked mental retardation/ Aarskog–Scott syndrome† |
Mental defect/short stature, hypertelorism, shawl scrotum, brachydactyly | RhoGEF; possible role in stimulation of actin polymerization and neurite outgrowth |
| FMR2 | X-linked mental retardation (FRAXE type) | Mild to moderate mental defect | Regulator of transcription, possibly involved in long-term memory and enhanced long-term potentiation |
| FTSJ1 | X-linked mental retardation-9/44 | Nonspecific X-linked mental retardation | tRNA modification and RNA translation |
| GDI1 | X-linked mental retardation-9/41 | Mild to moderate mental defect | Regulation of Rab4 and Rab5 pools, probably involved in the maturation of synaptic vesicles |
| IL1RAPL | X-linked mental retardation-21/34 | Moderate to severe mental defect | Regulator of dense-core-granule exocytosis, possible modulator of neurotransmitter releasing |
| JARID1C* | X-linked mental retardation (Claes-Jensen type) | Microcephaly, recurrent spasticity, epilepsy, short stature, facial anomalies, mental defect | Transcription factor; chromatin remodeling |
| MECP2* | Rett syndrome/X-linked mental retardation (Lubs type) | Neurodevelopmental regression, epilepsy, hand stereotypies, recurrent spasticity, recurrent spasticity/ mental defect | Silencing of transcription in neuronal genes |
| NLGN4 | X-linked mental retardation | Mild to moderate mental defect | Post-synaptic membrane protein that is involved in induction of presynaptic structures; linked to NMDA-type glutamatergic receptors |
| PQBP1* | Renpenning syndrome | Microcephaly, short stature, small testes, dysmorphic face, ocular colobomas, cardiac malformations, cleft palate | Polyglutamine-binding; mRNA splicing |
| RSK2* | X-linked mental retardation-19/ Coffin–Lowry syndrome |
Mild to moderate mental defect/ skeletal malformations, growth retardation, hearing deficit, paroxysmal movement disorders, cognitive impairment | Serine-threonine protein kinase; CREB phosphorylation; long-term memory |
| SLC6A8* | Cerebral creatine deficiency syndrome 1 | Epilepsy, mental retardation, severe speech delay, behavioral abnormalities, seizures, facial anomalies | Creatine transporter, maintenance of creatine pool in brain |
| TM4SF2 | X-linked mental retardation-58 | Mild to moderate mental defect | Modulation of integrin-mediated signaling, neurite outgrowth, possible role in synapse formation |
| XNP* | X-linked epilepsy, with variable learning disabilities and behavior disorders | Epilepsy, learning difficulties, macrocephaly, aggressive behavior, skeletal, urogenital and facial anomalles, α-thalassemia, short stature, spastic diplegia | DNA helicase; chromatin remodeling, DNA methylation and regulation of gene expression; regulator of cortical size |
| ZNF41 | X-linked mental retardation | Severe mental defect | Transcriptional regulator that is involved in chromatin remodeling |
*Mutated in both syndromic and non-syndromic forms of XLID; NMDA, N-methyl-D-aspartate.
Table 2. Genes responsible for non-syndromic form of X-linked intellectual disability.
ARX gene: Structure and function
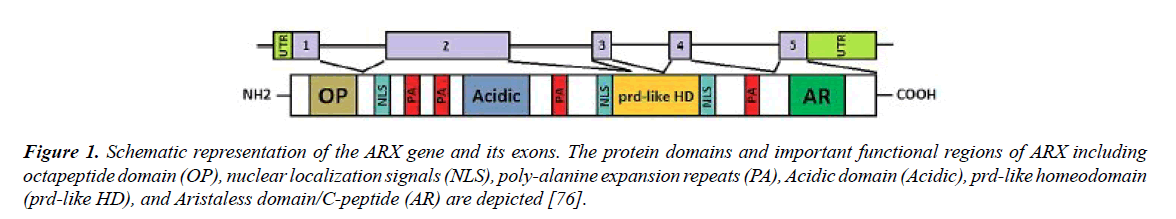
The Aristaless-related homeobox gene (ARX) is located on the Xp22.13. It consists of 5 exons (Figure 1) and is transcribed into 2.8kb mRNA. ARX mRNA encodes a 562 amino acids protein that seems to be a transcriptional activator and repressor with the main task in fetal and adult forebrain, pancreas and testis growth [45-49]. The expression of this gene is much more in fetal brain and has a critical role in differentiation and maintenance of neuronal cell during embryogenesis [47,50,51]. The structure of ARX protein is consists of some different compartments, including:
Figure 1. Schematic representation of the ARX gene and its exons. The protein domains and important functional regions of ARX including octapeptide domain (OP), nuclear localization signals (NLS), poly-alanine expansion repeats (PA), Acidic domain (Acidic), prd-like homeodomain (prd-like HD), and Aristaless domain/C-peptide (AR) are depicted [76].
(1) A highly conserved homeobox domain (repressor domain) (Table 3) that spans from amino acids 328 to 387. This part of protein is directly binds to DNA.
10 20 30 40 50 60 |
70 80 90 100 110 120 |
130 140 150 160 170 180 |
190 200 210 220 230 240 |
250 260 270 280 290 300 |
310 320 330 340 350 360 |
370 380 390 400 410 420 |
430 440 450 460 470 480 |
490 500 510 520 530 540 |
550 560 |
Table 3. Homeobox protein ARX, Homo sapiens, Q96QS3 [328-387].
(2) C-terminal OAR or aristaless domain (activator domain) which spans from amino acids 530 to 543 of protein (Figure 1).
(3) Octapeptid domain which is a receptor site beside the N-terminal of the ARX protein for some enhancer proteins that contribute to ARX functional activity adjustments.
(4) Four polyalanine tracts which are located between Hundredth-degree amino acid and 115, 144 and 155, 278 and 281, also 432 and 440, that each one has 16, 12, 7, and 9 residues respectively [52-54].
The 24 bp duplication in exon 2 is the most frequent mutation that found in this gene which is consists of 45% of all mutations. This mutations lead to an expansion in the second polyalanine tract of ARX protein from 12 to 20 residues [55-58]. Furthermore, addition of alanine residues to the first polyalanine tract is also reported [45,59]. These mutations influence the transcriptional repression traits of ARX protein [60]. The 4th tract is more conservative in comparison to others mentioned previously [61]. An acidic domain and three nuclear localization sequence motifs (NLS1, NLS2 and NLS3) are other parts of this protein [62-64]. It was determined that ARX gene is evolutionarily conserved in different species and according to Figures 2-4, the local similarity to its target binding sites is high. Also, the sequence alignment of this protein with other spices (Figure 2) confirms that the functional domains of ARX protein are highly conserved, thus it has been predicted that the mutations of this gene is highly pathogenic [65].
ARX and the frequency of its mutation
According to European XLID consortium, mutations of ARX gene has been found in 9.5% of families with X-linked intellectual disability and 7.5% of large families with 2 or more affected males from multi generations that are related with each other through an obligate carrier female [51,66-68].
Near to 70% of the families linked to Xp22.1 have the c.428_451dup (24 bp) which results in the expansion of the second poly (A) tract in the ARX protein and may lead to syndromic and non-syndromic forms of XLID [69-71]. The c.428_451dup with another known missense mutation in the exon 1 of ARX gene, c.98T>C (Leu33Pro) at octapeptid domain, are the main reasons of NS-XLID caused by ARX gene defects [72,73]. According to the study of S. S. Abedini, the later mutation accounts for 6.25% of the cases in the linked families from Iranian population, which is in agreement with the other published studies performed on different races [74].
ARX-associated phenotypes
ARX incapacitate mutations through exons and introns and subsequently different domains, are associated with a wide spectrum of phenotypes ranging from severe developmental abnormalities of the brain to syndromic forms of XLID. Early infantile epileptic encephalopathy-type 1 (OMIM#308350); Lissencephaly-type 2 (OMIM#300215); Hydranencephaly with abnormal genitalia(OMIM#300215);Proud Syndrome (OMIM#300004); Partington Syndrome (OMIM# 309510); X-linked Mental retardation, and ARX-related (OMIM#300419) are the known various syndromic phenotypes associated with ARX mutations [75].
How different mutations in this single transcription factor can produce different phenotypes is not completely understood. During the recent theory of Il-Taeg Cho and his colleagues, the ARX genes has interaction with different cofactors/transcription factors and regulate single target genes in different cell types.
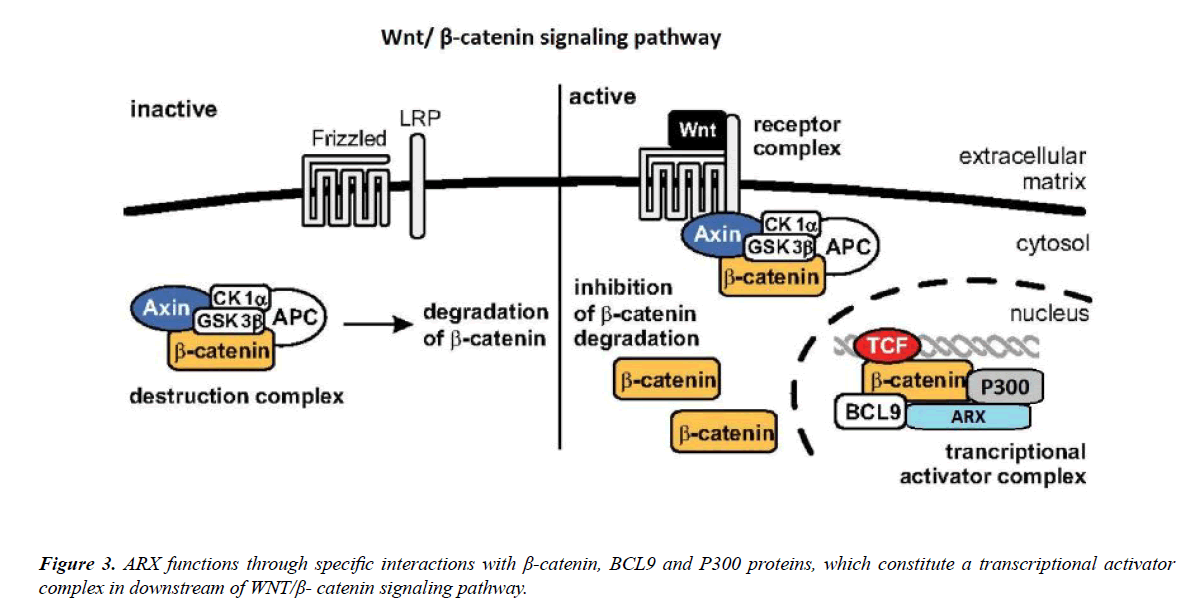
According to Il-Taeg Cho’s paper, by using the proteomics method, it was determined that the Wnt/β-catenin signaling pathway includes three components such as: B-cell CLL/ lymphoma 9 (BCL9), β-catenin (CTNNB1) and leucine-rich repeat flightless interacting protein 2 (LRRFIP2). They showed that ARX positively controls Wnt/β-catenin signaling and that the C-terminal domain of ARX interacts with the armadillo repeats of β-catenin to move forward Wnt/β-catenin signaling.
Furthermore, they understood that P300 and BCL9 also interact with ARX to adjust Wnt/β-catenin signaling. These data offer new insights into how ARX can exclusively regulate cortical neurogenesis, and link the role of ARX with Wnt/β-catenin signaling [76] (Figure 3).
Bioinformatics analysis
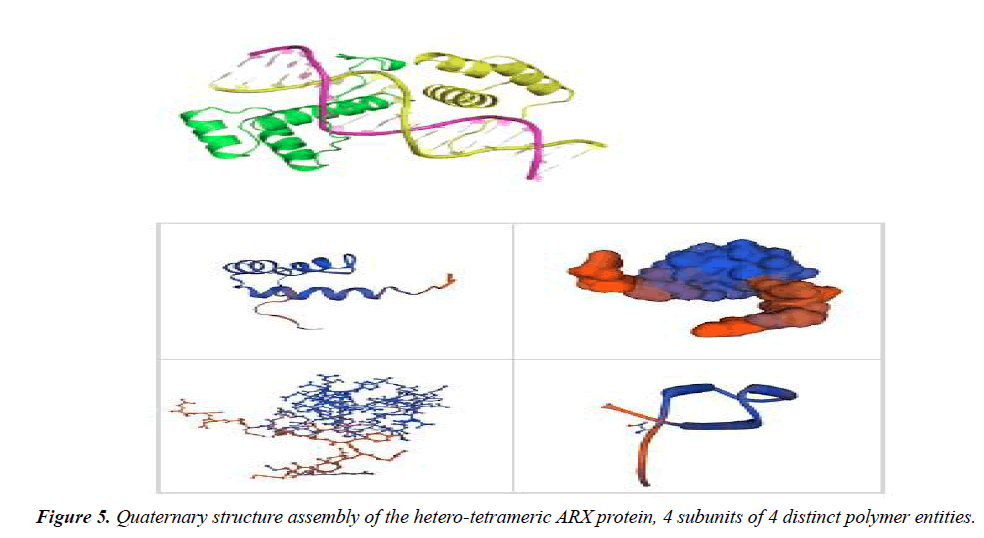
To study the molecular features, second (Table 4) and quaternary (Figure 5) structures of ARX gene, bioinformatics analysis was performed by using ExPASy software and SWISS-MODEL server, respectively. The phylogeny tree of ARX protein was also drawn using the software.
| Helix | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chain C: 5'-D(*GP*GP*CP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*GP*CP*GP*G)-3' 1 G G C T T A A T T A A T T G C G G |
||||||||||||||||||
| Chain D: 5'-D(*CP*CP*GP*CP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*GP*CP*C)-3' 1 C C G C A A T T A A T T A A G C C |
||||||||||||||||||
| Chain G: 5'-D(*GP*GP*CP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*GP*CP*GP*G)-3' 1 G G C T T A A T T A A T T G C G G |
||||||||||||||||||
| Chain H: 5'-D(*CP*CP*GP*CP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*GP*CP*C)-3' 1 C C G C A A T T A A T T A A G C C |
||||||||||||||||||
Table 4. Secondary structure of Homo sapiens ARX protein.
Discussion
As illustrated in Figure 1 and 2, it was determined that ARX gene is evolutionarily conserved in different species. Moreover, the sequence alignment of this protein with other spices confirms that the functional domains of ARX protein are highly conserved, and therefore it has been predicted that the mutations of this gene can be highly pathogenic.
Conclusion
Taken together, ARX is a highly conserved protein with a substantial role in an important developmental pathway and its deficiency can cause irreversible defects, mainly in brain, that leads to the development of XLID as a common form of intellectual disability.
References
- Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6(1):46-57.
- Leonard H, Wen X. The epidemiology of mental retardation: Challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002;8(3):117-34.
- Saraceno B. Atlas: Child and adolescent mental health resources: Global concerns, implications for the future. World Health Organization. 2005.
- Durkin MS, Khan NZ, Davidson LL, et al. Prenatal and postnatal risk factors for mental retardation among children in Bangladesh. Am J Epidemiol. 2000;152(11):1024-33.
- Durkin MS, Hasan ZM, Hasan KZ. Prevalence and correlates of mental retardation among children in Karachi, Pakistan. Am J Epidemiol. 1998;147(3):281-88.
- Durkin M. The epidemiology of developmental disabilities in low-income countries. Ment Retard Dev Disabil Res Rev. 2002;8(3):206-11.
- Costeff H, Cohen BE, Weller L. Parental consanguinity among Israeli mental retardates. Acta Paediatr Scand. 1972;61(4):452-8.
- Bashi J. Effects of inbreeding on cognitive performance. Nature. 1977;266(5601):440-2.
- Thompson BL, Levitt P, Stanwood GD. Prenatal exposure to drugs: Effects on brain development and implications for policy and education. Nature Rev Neurosci. 2009;10(4):303-12.
- Chaney RH, Givens CA, Watkins GP, et al. Birth injury as the cause of mental retardation. Obstet Gynecol. 1986;67(6):771-5.
- King BH, Toth KE, Hodapp RM, et al. Intellectual disability. In: Sadock BJ, Sadock VA, Ruiz P. Comprehensive textbook of psychiatry. Lippincott Williams & Wilkins, Philadelphia 1992;pp:3444-74.
- Liana K, Ayub M, Vincent JB. The genetic basis of non-syndromic intellectual disability: A review. J Neurodev Disord. 2010;2(4):182-209.
- Amélie P, Redin C, Mandel JL. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet. 2013;93(2):368-83.
- Lucy RF, Tarpey P. The genetics of mental retardation. Hum Mol Genet. 2006;15(2):R110-6.
- Bassani S, Zapata J, Gerosa L et al. The neurobiology of X-linked intellectual disability. Neuroscientist. 2013;19(5):541-2.
- Stevenson RE, Schwartz CE. Clinical and molecular contributions to the understanding of X-linked mental retardation. Cytogenet Genome Res. 2002;99(1-4):265-75.
- Penrose LS. Clinical and genetic study of 1280 cases of mental defect. London: HMSO, Lomdon, United Kingdom. 1938;p:159.
- Drillien CM. The incidence of mental and physical handicaps in school age children of very low birth weight. II. Pediatrics. 1967;39(2):238-47.
- McLaren J, Bryson SE. Review of recent epidemiological studies of mental retardation: Prevalence, associated disorders, and etiology. Am J Ment Retard. 1987;92(3):243-54.
- Baird PA, Sadovnick AD. Mental retardation in over half‐a‐million consecutive livebirths: An epidemiological study. Am J Ment Defic. 1985;89(4):323-30.
- Stevenson RE, Schwartz C, Schroer R. Mental retardation in South Carolina. I. Characteristics of the study population. Proc Greenwood Genet Center. 1996;50:176-83.
- Stoller A. Epidemiology of mental deficiency in Victoria. In: JD Van Zelt, ed. Proceedings of the Fourth Interstate Conference on Mental Deficiency. Melbourne, Australia: Australian Group for the Scientific Study of Mental Deficiency. 1965;pp:18-28.
- Hogan A. Visualizing carrier status: Fragile X sybndrome and genetic diagnosis since the 1940s. Endeavour. 2012;36(2):77-84.
- Jacquemont S, Des-Portes V, Hagerman R. Fragile X and X-linked mental retardation. In: Butler MG, Meaney FJ, editors. Genetics of Developmental Disabilities 1. Taylor & Francis; Boca Raton. 2005;pp:247-78.
- Hu H, Haas SA, Chelly J, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry. 2016;21(1):133-48.
- Lubs HA, Stevenson RE, Schwartz CE. Fragile X and X-linked intellectual disability: Four decades of discovery. Am J Hum Genet. 2012;90(4):579-90.
- Moore SJ, Strain L, Cole GF, et al. Fragile X syndrome with FMR1 and FMR2 deletion. J Med Genet. 1999;36(7):565-6.
- Bassell GJ, Warren ST. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201-14.
- Verkerk AJMH, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a [CGG] repeat coincident with a break point cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905-14.
- Crawford DC, Acuna JM, Sherman SL. FMR1 and the fragile X syndrome: Human genome epidemiology review. Genet Med. 2001;3(5):359-71.
- Knight SJ, Ritchie RJ, Chakrabarti L, et al. A study of FRAXE in mentally retarded individuals referred for fragile X syndrome (FRAXA) testing in the United Kingdom. Am J Hum Genet. 1996;58(5):906-13.
- Gecz J, Gedeon AK, Sutherland GR, et al. Identification of the gene FMR2, associated with FRAXE mental retardation. Nature Genet. 1996;13(1):105-8.
- Shen Y, Gibbs RA, Nelson DL. Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nature Genet. 1996;13(1):109-13.
- Gu Y, McIlwain KL, Weeber EJ, et al. Impaired conditioned fear and enhanced long-term potentiation in Fmr2 knock-out mice. J Neurosci. 2002;22(7):2753-63.
- Chelly J, Mandel JL. Monogenic causes of X-linked mental retardation. Nature Rev Genet. 2002;2(9):669-80.
- Chelly J. Breakthroughs in molecular and cellular mechanisms underlying X-linked mental retardation. Hum Mol Genet. 1999;8(10):1833-8.
- Ramakers GJ. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25(4):191-99.
- Billuart P, Bienvenu T, Ronce N, et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392(6679):923-26.
- Kutsche K, Yntema H, Brandt A, et al. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nature Genet. 2000;26(2):247-50.
- Pasteris NG, Cadle A, Logie LJ, et al. Isolation and analysis of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: A putative, rho/rac guanine nucleotide exchange factor. Cell. 1994;79:669-78.
- Govek EE, Newey SE, Akerman CJ, et al. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nature Neurosci. 2004;7(4):364-72.
- Allen KM, Gleeson JG, Bagrodia S, et al. PAK3 mutation in nonsyndromic X-linked mental retardation. Nature Genet. 1998;20(1):25-30.
- D'Adamo P, Menegon A, Nigro CL, et al. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nature Genet. 1998;19(2):134-9.
- Kulkarni G, Ranade S. Molecular studies on ARX gene in syndromic and non-syndromic mental retardation. Mol Cytogenet. 2014;7(S1):75.
- Bienvenu T, Poirier K, Friocourt G, et al. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet. 2002;11:981-91.
- Chiurazzi P, Tabolacci E, Neri G. X-Linked Mental Retardation (XLMR): From clinical conditions to cloned genes. Crit Rev Cl Lab Sci. 2004;41:117-58.
- Ohira R, Zhang YH, Guo W, et al. Human ARX gene: Genomic characterization and expression. Mol Genet Met. 2002;77:179-88.
- Poirier K, Esch HV, Friocourt G, et al. Neuroanatomical distribution of ARX in brain and its localisation in GABA ergic neurons. Brain Res Mol Brain Res. 2004;122(1):35-46.
- Kato M, Dobyns WB. X-linked lissencephaly with abnormal genitalia as a tangential migration disorder causing intractable epilepsy: proposal for a new term, “interneuronopathy”. J Child Neurol. 2005;20(4):392-7.
- Friocourt G, Poirier K, Rakic S, et al. The role of ARX in cortical development. Eur J Neurosci. 2006;23:869-76.
- Wigle JT, Eisenstat DD. Homeobox genes in vertebrate forebrain development and disease. Clin Genet. 2008;73(3):212-26.
- Strømme P, Mangelsdorf ME, Shaw MA, et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation. Nat Genet. 2002;30:441-5.
- Kitamura K, Yanazawa M, Sugiyama N, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359-69.
- Kato M, Das S, Petras K, et al. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum Mutat. 2004;23:147-59.
- Turner G, Partington M, Kerr B, et al. Variable expression of mental retardation, autism, seizures, and dystonic hand movements in two families with an identical ARX gene mutation. Am J Med Genet. 2002;112:405-11.
- Frints SGM, Froyen G, Marynen P, et al. Re-evaluation of MRX36 family after discovery of an ARX gene mutation reveals mild neurological features of Partington syndrome. Am J Med Genet. 2002;112:427-8.
- Grønskov K, Hjalgrim H, Nielsen IM, et al. Screening of the ARX gene in 682 retarded males. Eur J Hum Genet. 2004;12:701-5.
- Reish O, Fullston T, Regev M, et al. A novel de novo 27 bp duplication of the ARX gene, resulting from postzygotic mosaicism and leading to three severely affected males in two generations. Am J Med Genet A. 2008;149(8):1655-60.
- Shinozaki Y, Osawa M, Sakuma H, et al. Expansion of the first polyalanine tract of the ARX gene in a boy presenting with generalized dystonia in the absence of infantile spasms. Brain Dev. 2009;31(6):469-72.
- Nasrallah MP, Cho G, Simonet JC, et al. Differential effects of a polyalanine tract expansion in Arx on neural development and gene expression. Hum Mol Genet. 2011;21(5):1090-8.
- McKenzie O, Ponte I, Mangelsdorf M, et al. Aristaless-related homeobox gene, the gene responsible for West syndrome and related disorders, is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience. 2007;146(1):236-47.
- Cho G, Nasrallah MP, Lim Y, et al. Distinct DNA binding and transcriptional repression characteristics related to different ARX mutations. Neurogenetics. 2012;13(1):23-9.
- Lin W, Ye W, Cai L, et al. The roles of multiple importins for nuclear import of murine aristaless-related homeobox protein. Journal of Biological Chemistry. 2008;284(30):20428-39.
- Shoubridge C, Tan MH, Fullston T, et al. Mutations in the nuclear localization sequence of the Aristaless related homeobox; sequestration of mutant ARX with IPO13 disrupts normal subcellular distribution of the transcription factor and retards cell division. Pathogenetics. 2010;3(1).
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny. fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465-9.
- Takahashi T, Fukuyama Y. Biology of seizures susceptibility in development brain. Medical. 2008;p:66.
- Cossée M, Faivre L, Philippe C, et al. ARX polyalanine expansions are highly implicated in familial cases of mental retardation with infantile epilepsy and/or hand dystonia. Am J Med Genet A. 2011;155(1):98-105.
- Brouwer APM, Yntema HG, Kleefstra T, et al. Mutation frequencies of X‐linked mental retardation genes in families from the EuroMRX consortium. Hum Mutat. 2002;28(2):207-8.
- Poirier K, Lacombe D, Dussardier BG, et al. Screening of ARX in mental retardation families: consequences for the strategy of molecular diagnosis. Neurogenetics. 2006;7(1):39-46.
- Mandel JL, Chelly J. Monogenic X-linked mental retardation: Is it as frequent as currently estimated? The paradox of the ARX (Aristaless X) mutations. Eur J Hum Genet. 2004;12(9):689-93.
- Shoubridge C, Fullston T, Gécz J. ARX spectrum disorders: making inroads into the molecular pathology. Hum Mutat. 2010;31(8):889-900.
- Bienvenu T. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet. 2002;11(8):981-91.
- Abedini SS, Kahrizi K, Behjati F, et al. Mutational screening of ARX gene in Iranian families with X-linked intellectual disability. Arch Iran Med. 2012;15(6):361-5.
- Cho TI, Lim Y, Jeffrey AG, et al. Aristaless related homeobox (ARX) interacts with β-Catenin, BCL9, and P300 to regulate canonical wnt signaling. PloS one. 2017;12(1):e0170282.
- Gage BK, Asadi A, Baker RK, et al. The role of ARX in human pancreatic endocrine specification. PloS one. 2015;10(12):e0144100.
- Grossmann TN, Yeh JTH, Bowman BR, et al. Inhibition of oncogenic Wnt signaling through direct targeting of β-catenin. Proc Natl Acad Sci. 2012;109(44):17942-7.