- Journal of RNA and Genomics (2014) Volume 10, Issue 1
Aptamer-mediated selective delivery of short RNA therapeutics in cancer cells
| Carla Lucia Esposito, Silvia Catuogno and Vittorio de Franciscis* Istituto di Endocrinologia ed Oncologia Sperimentale, CNR, Naples, Italy |
| *Correspondence to:Vittorio de Franciscis, Email: defranci@unina.it, Tel: +39 0813722343 |
| Received: 05 September 2014; Accepted: 24 September 2014; Published: 03 October 2014 |
| © Copyright The Author(s). First Published by Library Publishing Media. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License p://creativecommons.org/licenses/ by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details. |
Abstract
RNA interference (RNAi) is an important biological process that ultimately leads to suppression of gene expression. Activators of RNAi are typically small interfering RNAs (siRNA) and microRNAs (miRNA) that offer considerable therapeutic potnetial. However, a major obstacle to take these these molecules to the clinic is the absence of safe and reliable means for their specific delivery to target cells. In this regard, a highly promising class of molecules is represented by nucleic acid aptamers. These are short, structured, single-stranded RNAs or DNAs oligonucleotides that, by binding with high specificity to target molecules, provide high affinity ligands and potential antagonists of disease-associated proteins. Further, because of the high binding specificity, aptamers represent a powerful tool for the selective delivery of therapeutic cargos, including mi/ siRNAs, chemotherapeutics, toxins and nanoparticles to cancer cells or tissues, thus potentially increasing the efficacy of a given therapy as well as reducing toxicity. In this review, we will focus on recent advances in the field of aptamer-mediated mi/siRNA delivery, discussing their potential and challenges in cancer therapy.
KEYWORDS |
| microRNA, siRNA, aptamer, targeted delivery, cancer |
INTRODUCTION |
| Therapeutic molecules that specifically target cancer cells are the key to the development of patient-tailored cancer treatment. The selective targeting of cancer cells would enable treatments that inhibit tumor growth and invasion while sparing the tumor surrounding normal cells, thus reducing the adverse side effects frequently caused by conventional chemotherapies (Toniatti et al, 2014). |
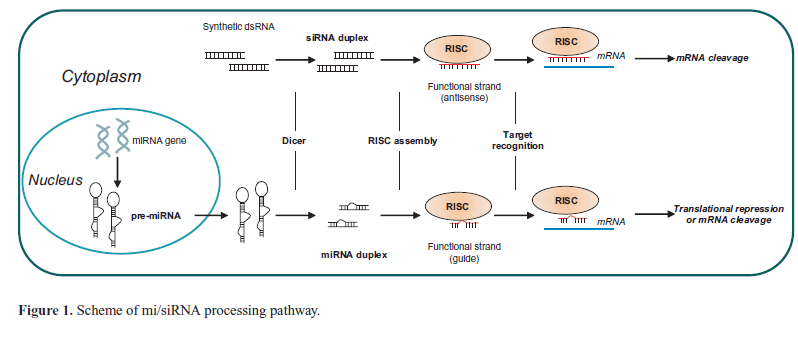
| Because of the versatility of the RNAi process, RNA-based gene therapy is emerging as a potential effective and safe approach to target various human diseases, including cancers (Uchino et al, 2013). RNAi molecules include both extrinsic short interfering RNAs (siRNAs) and intrinsic antisense RNAs (AS-RNA), and microRNAs (miRNAs), and modulate post-transcriptional and sequence-specific gene silencing. Si/miRNAs are processed by the ribonuclease DICER to produce short double-stranded RNAs (dsRNAs) of 20-24 base pairs that in the cytoplasm are recognized by the RNA-induced silencing complex (RISC). The RISC drives the strand that directs silencing (guide strand) to the target mRNA, while the other strand (passenger strand) is degraded. Depending on the degree of complementarity, the hybridization of guide strand to the mRNA prevents translation or induces degradation (Rand et al, 2005). SiRNAs that show a perfect complementarity to their target mRNA, induce gene silencing through a sequence-specific cleavage of the target RNA, whereas microRNAs, that show a partial complementarity, can mediate translational repression or transcript degradation (Figure 1). |
| Using RNAi to inhibit the action of genes relevant for cancer cell survival and progression presents several advantages over other inhibitors as small molecules, peptides and antibodies, since the elevated target specificity is coupled to low toxicity. Chemical modifications can be easily introduced in sequence to increase resistance to nucleases, stability or binding properties (Shukla et al, 2010). In addition, |
 |
| solid-phase synthesis of short nucleic acids has made the large-scale production cost-effective. To-date numerous pre-clinical studies have provided encouraging results for use of this class of molecules as cancer therapeutics. While the RNAi technology offer potential to inhibit expression of specific and multiple target genes, raising the hope of patient tailored therapies; however, several challenges hinder their successful translation into the clinic. A major obstacle is represented by the necessity to develop safe and effective means for their delivery, enabling the selective accumulation of RNAi at target sites, their intracellular uptake, correct processing and functional silencing of target genes. |
| In this regard, nucleic acid aptamers are emerging as very promising molecules. Aptamers are short, structured, single-stranded RNA or DNA ligands that bind to target molecules with high specificity and affinity (Kd in the low nanomolar-picomolar range). In the last decade, aptamers that target the extracellular domain of transmembrane receptors overexpressed in tumors have been generated, thus becoming, along with monoclonal antibodies, ideal tools for the specific recognition of cancer cell surface (Cerchia and de Franciscis, 2010). Thanks to their unique characteristics, cancer targeting aptamers are revealing a promising mean to selectively drive nanoparticle-based cargoes to affected cancer tissues. Further, because of the chemical versatility of nucleic acids, aptamers have been as well directly conjugated to different secondary reagents such as chemotherapeutics, imaging probes and siRNAs/ miRNAs to be selectively driven toward the target cells. |
| In this review, we will discuss the various approaches adopted to use aptamers for the selective delivery of si/ miRNAs to cancer cells with a special focus on the design of aptamer-RNAi conjugates. |
APTAMERS AS DELIVERY TOOLS |
| Aptamers are generated from high complexity pools through a combinatorial process named Systematic Evolution of Ligands by EXponential enrichment (SELEX) to tightly bind to their proper targets (Tuek and Gold, 1990). Various aptamers have been raised that bind to and inhibit the function of proteins involved in cancer. As such, the aptamer technology is now emerging for its potential as an effective diagnostic and therapeutic tool, and many aptamers are currently in clinical and preclinical trials (Esposito et al, 2011). |
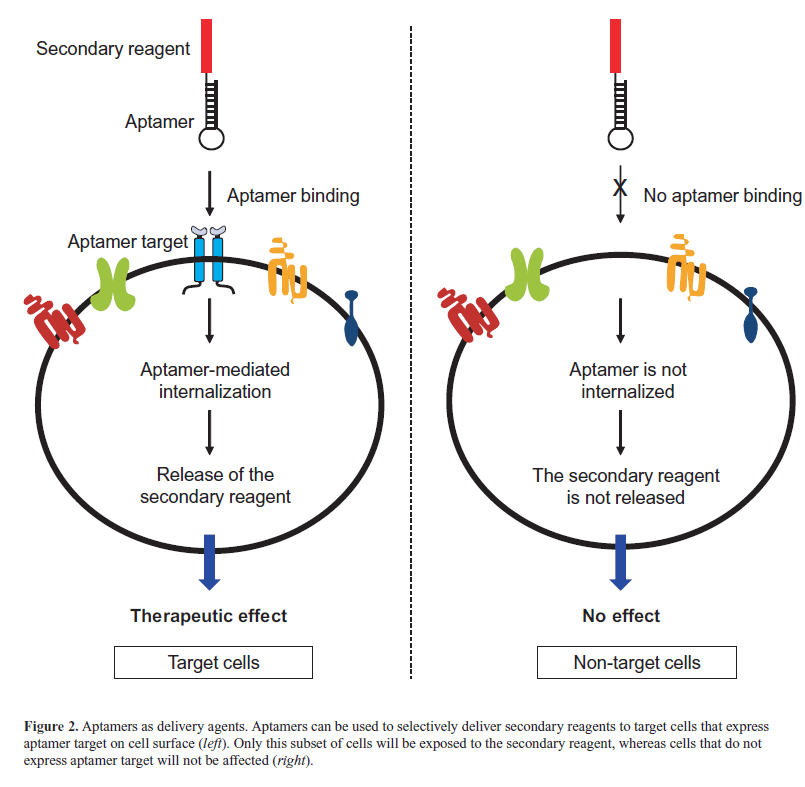
| Aptamers were first identified as cancer therapeutics because of their ability to inhibit their targets and for their appropriate pharmacokinetic and pharmacodynamic properties. More recently, thanks to their excellent targeting properties, they have been developed as delivery tools for secondary therapeutic agents and nanoparticles. To-date, they have been successfully conjugated to different active molecules, including nanoparticles (Farokhzad et al, 2006; Wang et al, 2013), anti-cancer therapeutics (Bagalkot et al, 2006), toxins (Chu et al, 2006a), enzymes (Chen et al, 2008), radionuclides (Hicke et al, 2006), siRNAs (Zhou and Rossi, 2010) and miRNAs (Wu et al, 2011; Dai et al, 2012; Liu et al, 2012; Esposito et al, 2014). In these conjugates, the aptamer moiety is used to selectively drive the secondary reagent to affected tissues or cells that overexpress the aptamer target, eluding the accumulation of drugs in nontarget cells. Consequently, the efficacy of the therapy is increased and the possibility of unwanted side effects is reduced (Figure 2). The use of aptamers as delivery tools is particularly relevant in the case of conjugates with si/ miRNA therapeutics (Wu et al, 2014). |
| Aptamers show important advantages over viral and lipid nanoparticle-based approaches proposed for iRNA delivery. Even if adenovirus- and adeno-associated viruses have shown high efficacy, they give problems of insertional mutagenesis and immunogenicity (Kay, 2011). On the other hand, nanoparticle-based systems have achieved a limited effectiveness in vivo, resulting in a preferentially accumulation in the liver, a dose-dependent toxicity and hepatotoxicity (Zhou et al, 2013a). Thus the use of aptamers offers the possibility to overcome these problems enabling the specific accumulation of siRNAs in target tumor cells in a safe and effective manner. |
| Aptamer-siRNA delivery |
| To-date, the use of synthetic siRNAs has been largely adopted to suppress the expression of pathologically important genes in |
 |
| cancer and several preclinical studies have shown the effective inhibition of tumor cell growth, angiogenesis, metastasis and chemo-resistance (Bora et al, 2012; Rao et al, 2013). Given the unprecedented potential impact in using si/miRNAs for personalized cancer therapy, a great effort has been invested to develop approaches enabling their safe and specific delivery. In this respect, using aptamers as delivery moieties revealed as a safe and effective approach for the specific recognition of solid tumors in preclinical studies. Aptamers against the extracellular domain of the prostate-specific membrane antigens (PSMA), A9 and A10 aptamers (Lupold et al, 2002), have been extensively characterized for siRNA delivery by developing different approaches based either on non-covalent or covalent conjugation. |
| One of the first studies reported the development of a non-covalent approach that employed the use of a streptavidin connector (Figure 3A) (Chu et al, 2006b). The A9 aptamer and the siRNA targeting laminin A/C and GAPDH genes were modified with biotin and then assembled via a streptavidin bridge. The treatment of PSMA-positive LNCaP cells with the conjugates resulted in a gene expression inhibition comparable to those observed with conventional lipid reagents. |
| In the same year, McNamara et al (2006) described an elegant covalent approach in which the aptamer is extended at the 3’end with a tail complementary to the antisense strand of the siRNA (nonfunctional or sense strand) and then annealed with the siRNA antisense strand (functional strand, Figure 3B), generating a completely RNA-based molecule. This approach was used to conjugate the anti-PSMA A10 aptamer with siRNAs targeting survival genes, the polo-like kinase 1 (PLK1) or BCL-2. The resulting chimeras specifically silenced target genes and induced cell death in PSMA-positive cancer cells. In a later study, (Dassie et al, 2009) the A10-Plk1-siRNA chimera was further optimized for in vivo application by truncating the aptamer portion, swapping the sense and antisense strands of the siRNA |
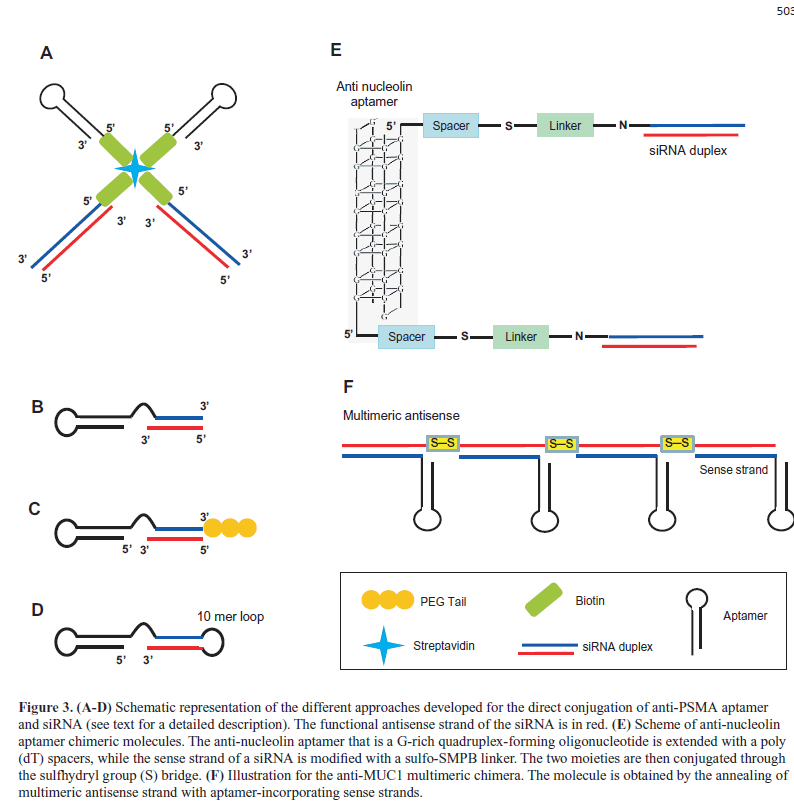 |
| portion and adding a two-nucleotide 3′-overhang and a PEG tail (Figure 3C). By suppressing the expression of the Plk-1 gene, the resulting conjugate effectively inhibited the growth of PSMA-positive tumors after systemic administration. |
| The A10 aptamer has been further covalently conjugated to different siRNAs, including those targeting the eukaryotic elongation factor-2 (Wullner U et al, 2008) or two key components of the nonsense-mediated mRNA decay (Pastor et al, 2010). In addition, a truncated version of A10 aptamer (A10-3) was linked to a short hairpin RNAs (shRNAs) against the DNA-activated protein kinase by generating a single molecules completely modified with 2’ fluoro-pyrimidine (2’-F-Py) (Ni X et al, 2011). In this chimera, the 3’-terminus of the aptamer was extended with the siRNA sense strand, followed by a 10-mer loop sequence and by the siRNA antisense strand (Figure 3D). The resulting molecules sensitized PSMA-positive cells to ionizing radiation therapy. |
| Several other aptamers against cell surface proteins overexpressed on cancer cells have been used for siRNA delivery. For example, the aptamer against nucleolin AS1411 (Bates et al, 2009) was recently conjugated to siRNAs specific for snail family zinc finger 2 (SLUG) and neuropilin 1 (NRP1) proteins (Lai et al, 2014). In the chimeric molecules (aptNCL-SLUGsiR and aptNCL-NRP1siR), the aptamer portion was extended with a poly(dT) spacer and conjugated through the sulfhydryl group (S) bridge to the siRNA sense strand modified with a sulfo-SMPB linker (Figure 3E). AptNCL-SLUGsiR and aptNCL-NRP1siR specifically knocked down the expressions of SLUG and NRP1 in nucleolin-positive cancer cells and, when used in combination, significantly reduced cancer cell migration and invasion in vitro and inhibited tumor growth, invasiveness and angiogenesis in vivo. In addition, AS1411 aptamer was used as targeting agent on PEGylated cationic liposome containing Braf specific siRNA. The complex showed an efficient tumor- targeting capability and inhibited tumor growth in xenograft of A375 melanoma cells (Li et al, 2014). |
| Another interesting cancer-related surface target is the epidermal growth factor receptor (EGFR) and its activated deletion variant (EGFRvIII), the most common EGFR mutation found in many malignancies including glioma. By using the streptavidin-biotin approach a DNA aptamer against EGFRvIII (Tan et al, 2013) was used to deliver c-Met siRNA into glioma U87MG cells overepressing EGFRvIII (Zhang et al, 2014). The efficient delivery of c?Met siRNA resulted in increasing apoptosis and inhibiting proliferation of target cells. In addition, aptamers specific for Her-2 positive breast cancer cells were covalently conjugated to BCL-2 siRNA, generating a chimera able to sensitize cells to chemotherapy (Thiel et al, 2012). |
| By using a DNA aptamer against mucin 1 (MUC1), that is overexpressed in different tumors (Kufe, 2009), it has been recently proposed a promising approach in which chemically conjugated multimeric antisense strands were annealed with aptamer-guide strands (Figure 3F). Compared to conventional chimeras, this multimeric construct enhanced siRNA intracellular delivery (Yoo et al, 2014). |
| Aptamer-targeted delivery has been also applied in cancer immunotherapy. In order to enhance antitumor immunity, Berezhnoy et al (2014) conjugated an aptamer that binds the 4-1BB, a costimulatory molecule of CD8+ T cells, with a siRNA targeting Raptor, a component of the mammalian target of rapamycin complex 1 (mTORC1). They found that the systemic administration of the chimera downregulated mTORC1 activity in CD8+ T cells, leading to a potent memory response that potentiated antitumor immunity. In the same year, Herrmann et al (2014) covalently liked an aptamer against the cytotoxic T lymphocyte–associated antigen 4 (CTLA4), that is upregulated in tumor associated CD8+ T cells, to siRNA targeting STAT-3, that induce peripheral tolerance in tumor associated immune cells inhibiting CD8+ T cell activity. The silencing of STAT-3 by the chimera resulted in the activation of tumor antigen– specific T cells and reduced tumor growth and metastasis in different mouse tumor models. Thus the studies concerning the use of aptamers for the delivery of siRNAs are growing rapidly and strongly support the importance of these two class of molecules for cancer treatment. |
| Aptamer-miRNA delivery |
| Endogenous miRNAs regulate important biological process including cancer initiation, progression and metastasis, showing a great potential as cancer therapeutics. Deregulation of miRNA expression has been involved in different malignancies and it is ascertained that miRNAs may act as oncogenes (oncomirs) or tumor suppressors, depending on the cellular function inhibited (Kasinski and Slack, 2011; di Leva et al, 2013). |
| MiRNAs have the potential to target simultaneously multiple genes that cooperate in the same pathway, thus resulting in an efficient cell reprogramming. As such, it makes unquestionable the development of a safe mean for the specific delivery of “oncosuppressor” miRNAs enabling the reduction of off-target effects while increasing the therapeutic effectiveness. Therefore, given the progresses in the design of aptamer-based strategies for siRNA delivery, it has been recently explored the use of aptamers as delivery moieties for microRNAs. In a first study, a second generation aptamer against PSMA (A10-3.2) was conjugated to a polyamidoamine (PAMAM)-based microRNA (miR-15a and miR-16-1) using PEG as a spacer. The construct demonstrated to be able to selectively deliver the miRNA moiety into LNCaP (PSMA positive) prostate cancer cells, inducing cell death in vitro (Wu et al, 2011). The same aptamer was recently used as recognition ligand in an atelocollagen (ATE)-based microRNA (miRNA; miR- 15a and miR-16-1) vector to target prostate cancer bone metastasis (Hao et al, 2014). |
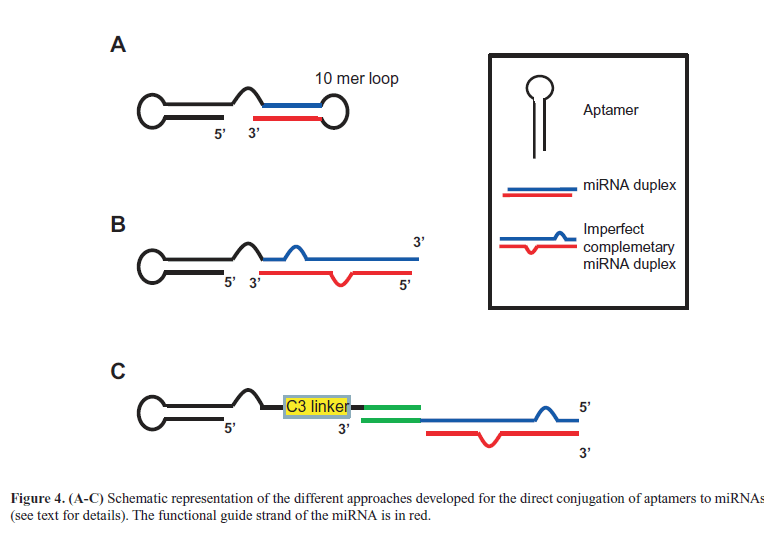
| Liu et al (2012) then described a direct combination between aptamers and miRNAs. The molecule, in which the anti-MUC1 aptamer was conjugated to let-7i miRNA, sensitized OVCAR-3 ovarian cancer cells to paclitaxel. The same aptamer was also combined to miR-29b that, by inhibiting DNA methyltransferases expression, is able to induce PTEN gene. Thus, the resulting chimera (Figure 4A) induced apoptosis in OVCAR-3 cells (Dai et al, 2012). More recently, we developed a novel multifunctional aptamer-miRNA conjugate (named GL21.T-let) (Esposito et al, 2014) by combining a previously characterized anti-Axl receptor inhibitory aptamer named GL21.T (Cerchia et al, 2012) with the tumor suppressor let-7g miRNA (Boyerinas et al, 2010). For the conjugation strategy, the aptamer was extended with the passanger strand of the miRNA and then annealed with the guide strand (Figure 4B). Notably, instead of using a perfect complementary 22-mer miRNA duplex, we introduced an internal partial complementarity and a longer sequence (26-27 bases) with the aim to have a better DICER substrate (Amarzguioui and Rossi, 2008). We demonstrated that GL21.T-let was selectively delivered to Axl-expressing cancer cells, processed by the iRNA machinery, leading to silence let-7g targets in vitro and in vivo. Importantly, the conjugate combined the miRNA activity with the aptamer function (Axl signaling inhibition), resulting in an effective inhibition of cell migration and survival in vitro and of tumor growth in vivo. Moreover, we also reported another conjugation strategies via a 17-mer “stick” complementary sequence (Figure 4C) that is based on rules already described for aptamer-siRNA chimeras for HIV treatment (Zhou et al, 2013b). The abnormal high expression of oncomir in cancer cells greatly contributes to the malignant phenotype. For an effective therapy, it is thus required to design single strand antisense oligonucleotides (named anti-miRs) able to antagonize the oncomir action. |
| To-date only a few studies are present in literature that describe the development of theragnostic nanoparticles (MFAS) containing AS1411 aptamer as targeting molecule and miR-221 molecular beacon (miR-221 MB) complementary to the oncogenic miRNA-221 (Kim et al, 2012). Once delivered to nucleolin positive cells, the miR-221 MB released from the MFAS was able to image miRNA-221 in the cytoplasm and simultaneouslyinhibiting the miRNA function. The selective delivery of anti-miRs to target cancer cells is still in its infancy; nevertheless, the development of aptamer-mediated approaches represent a concrete possibility to achieve this goal. |
 |
CONCLUSIONS |
| Gene therapies based on the use of iRNA to modulate gene expression are revealing as highly promising approaches for the treatment of canvers. Aptamers are emerging as safe and effective molecules enabling the specific recognition of target cells in vivo, the intracellular uptake and the functional processing of the RNAi, ultimately leading to proper target gene silencing. To-date, diverse approaches have been adopted to conjugate aptamers with siRNAs and, more recently, aptamer-microRNA chimeras have been described. Therefore, even if the aptamer-based anti-miRs delivery has been still poorly addressed, it represents an important challenge with broad applicability to treat many human diseases, including cancer, metabolic, cardiologic and infectious diseases. |
ACKNOWLEDGEMENTS |
| This work was supported by funds from: MIUR grant, MERIT RBNE08YFN3_001 (VdF); AIRC # 13345 (VdF); the Italian Ministry of Economy and Finance, Project FaReBio di Qualità (VdF); Grant CNR “Medicina Personalizzata” (VdF); Compagnia San Paolo # 2011.1172 (VdF); CNR Flagship Project NanoMax (DESIRED) 2012- 2014 (VdF); Italian Ministry of Health GR-2011-02352546 (CLE). |
COMPETING INTERESTS |
| None declared. |
LIST OF ABBREVIATIONS |
| iRNA; RNA interference |
| siRNA; short interfering RNAs |
| miRNAs; microRNAs |
| dsRNA; double-stranded RNA |
| RISC; RNA-induced silencing complex |
| SELEX; Systematic Evolution of Ligands by Exponential enrichment |
| PLK1; polo-like kinase 1 |
| shRNA; short hairpin RNA |
| 2’-F-Py; 2’ fluoro-pyrimidine |
| SLUG; Snail family zinc finger |
| NRP1; neuropilin 1 |
| MUC1; mucin 1 |
| mTORC1, rapamycin complex 1 |
References
|